Method of degrading lignin of plants with engineering bacteria of saccharomyces cerevisiae
A technology of Saccharomyces cerevisiae and lignin, applied in the field of construction and application of microbial engineering bacteria, which can solve the problems of low enzyme production, long cycle of degrading lignin, slow growth of white rot fungi, etc., and achieve the effect of low nutritional requirements and rapid growth and reproduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
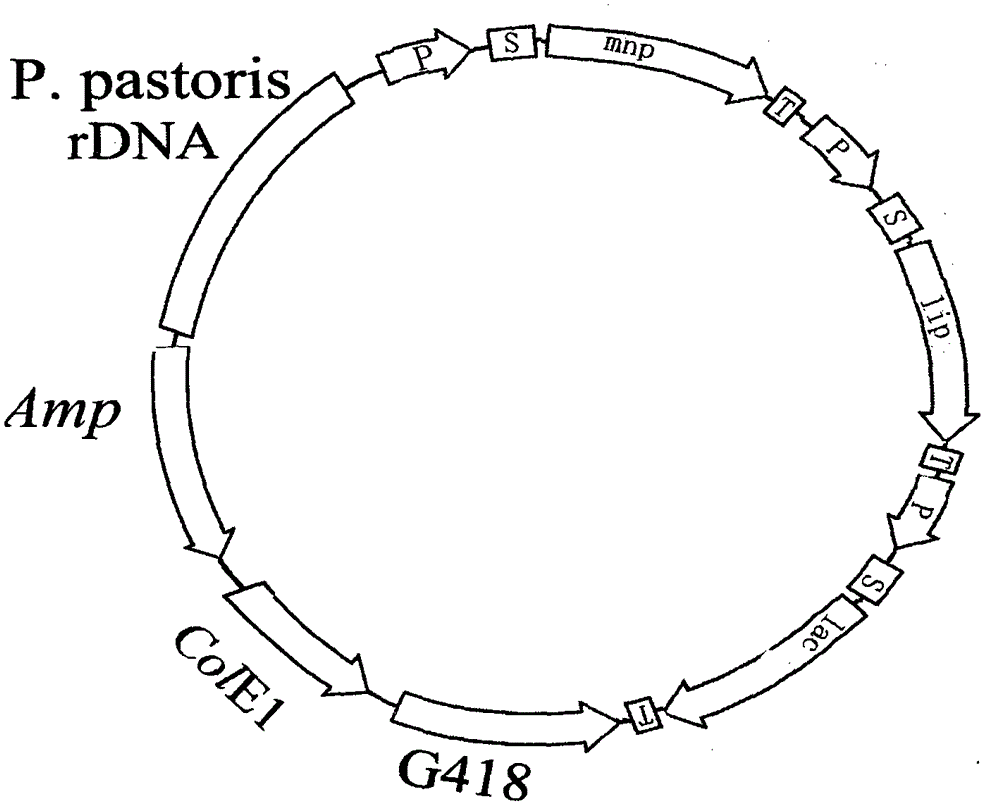
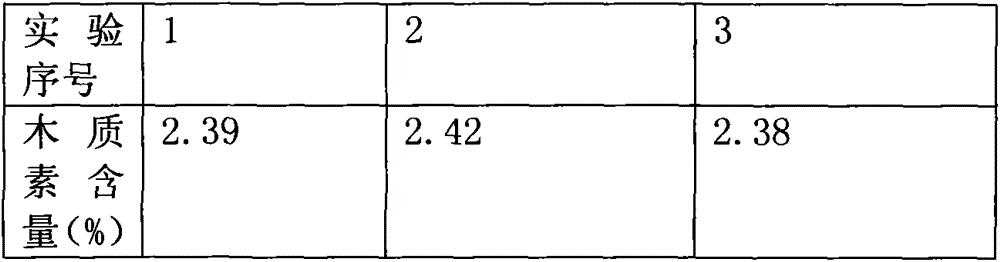
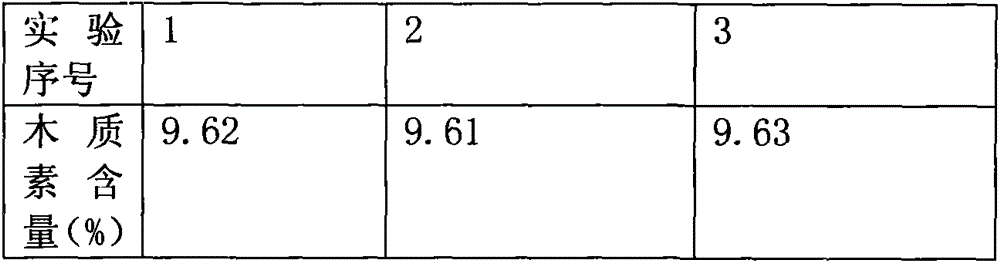
Image
Examples
Embodiment
[0018] 1. Obtain the relevant originals required for the enzyme gene and the construction of the expression vector
[0019] (1) PCR amplification of the promoter sequence of phosphoglyceraldehyde dehydrogenation of Saccharomyces cerevisiae
[0020] The cell wall of Saccharomyces cerevisiae was cleaved with helicase, the genomic DNA of Saccharomyces cerevisiae was extracted, and the following primers were used: 5'TCGAGTTTATCATTATCAAT3' and 5'TCGAAACTAAGTTCTTGGTG3') for PCR amplification, and the PCR product was sequenced and analyzed with BLAST software provided by NCBI to prove that it was Saccharomyces cerevisiae Promoter sequence of the yeast glyceraldehyde triphosphate dehydrogenase gene.
[0021] (2) PCR amplification of the laccase gene of Bacillus subtilis
[0022] The cell wall of Bacillus subtilis was cleaved with helicase, the genomic DNA of Bacillus subtilis was extracted, and the upstream primer (5AA CCTAGG ATGACACTTGAAAAATTTGTGGATGC3') and downstream primer (5'A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com