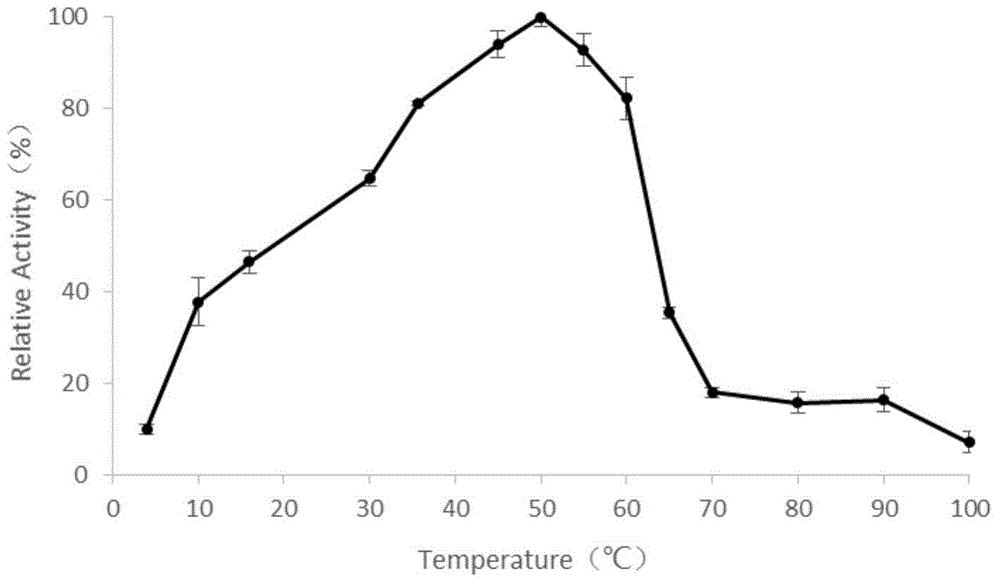
Novel alpha-amylase and application thereof
A technology of amylase and enzymatic properties, applied in the field of new α-amylase, to achieve the effect of wide temperature tolerance range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0012] Example 1: Acquisition of α-amylase gene laxh3357
[0013] Through the analysis of the whole genome of the new deep-sea bacteria XH031, three amylase genes and their gene sequences were obtained, and the upstream primer (5'-CGGAATTCATGCACCCTCGACCGGC-3') and the downstream primer (5'-CCCTCGAGACGCCGCCACATCCG -3') Using genomic DNA as a template, carry out PCR reaction, the PCR reaction composition is as follows (50 μ l reaction system): ddH 2 O 11.5 μl, upstream and downstream primers 0.5 μl each, DNA template 2 μl, 2×GC Buffer 25 μl, Taq 0.5 μl, dNTP Mixture 8 μl. The reaction conditions were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 65°C for 30 s, extension at 72°C for 3 min for 30 s, and final extension at 72°C for 10 min, a total of 30 cycles. After the reaction, the PCR product was recovered to obtain the α-amylase gene laxh3357. laxh3357 is one of the α-amylase genes, its nucleotide sequence is SEQ ID NO: 2, the encoded amino...
Embodiment 2
[0014] Example 2: Construction of Escherichia coli cloning vector PUCm-T-laxh3357.
[0015] The α-amylase gene laxh3357 was connected to the carrier PUCm-T by using DNA Ligation Kit. The connection system (10 μl) was as follows: SolutionI 5 μl, DNA fragment 4 μl, PUCm-T carrier 1 μl. The connection solution obtained by ligation at 16°C for 16 hours can be used to obtain the E. coli cloning vector PUCm-T-laxh3357, which is used to transform E. coli JM109.
Embodiment 3
[0016] Example 3: Construction of Escherichia coli recombinant strain JM109-PUCm-T-laxh3357
[0017] Add 200 μl of thawed competent Ecoli.JM109 and 10 μl of the connection solution obtained in Example 2 to a 2 ml Eppendorf tube, ice-bath for 30 minutes, heat shock at 42°C for 90 seconds, ice-bath for 15 minutes, add 800 μl of LB medium, and culture with shaking at 37°C for 45 minutes. The bacterial solution was mixed with 4 μl IPTG and 40 μl X-gal, spread on the LB plate containing 100 μg / ml ampicillin, and incubated at 37°C for 12-14h. Pick white colonies for PCR and double enzyme digestion detection, the enzyme digestion system (20μl) is as follows: ddH 2 O 8 μl, PUCm-T-laxh3357 plasmid DNA 8 μl, EcoRI 1 μl, XholI 1 μl, 10×H Buffer 2 μl, those with 1428 bp specific band in agarose gel electrophoresis were positive transformation clones, that is, Escherichia coli JM109- containing PUCm-T-laxh3357 PUCm-T-laxh3357. Send 1ml of the positive clone bacteria solution for testing,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com