Gene capable of keeping efficient nitrogen fixing capability of strain
A nitrogen fixation and gene technology, applied in the field of genetic engineering, can solve the problems of no microbial start codon and stop codon, and no transcription and translation of proteins have been found, so as to achieve the effect of high nitrogen fixation activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
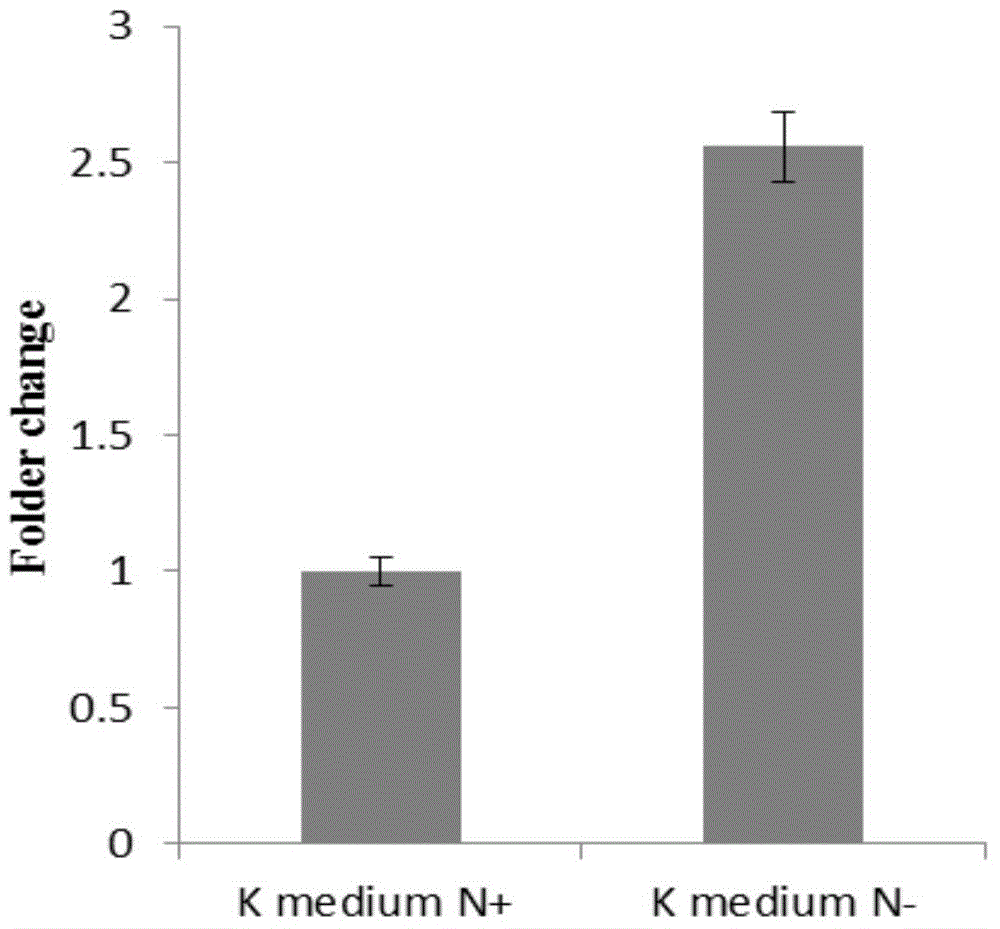
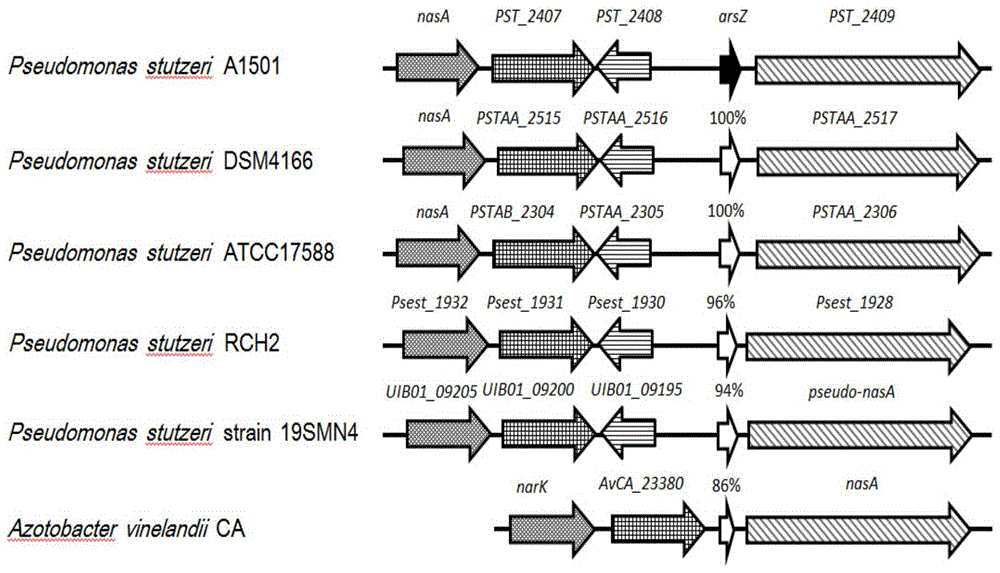
[0054] Example 1 Functional verification of arsZ gene expression specifically responding to environmental nitrogen signals
[0055] The experimental steps are as follows:
[0056] (1) Activate Pseudomonas stutzeri A1501 bacteria in LB liquid medium, and culture overnight at 30°C;
[0057] (2) Centrifuge the thallus at 4000rpm / 10min the next day, and wash the thallus twice with normal saline;
[0058] (3) Suspend the bacteria with physiological saline and adjust the OD 600 ≈1.0;
[0059] (4) Cultivate the bacteria in the following phases: nitrogen and aerobic conditions (CNO, K medium), nitrogen-free and aerobic conditions (CN - No nitrogen source was added to the O, K medium, and argon was filled to exhaust the air for three minutes, and then 0.5% oxygen was injected), and the OD was adjusted 600 ≈0.5;
[0060] (5) After shaking the culture solution at 30°C for 0.5h, centrifuge at 8000rpm for 5min to collect the bacteria;
[0061] (6) Total bacterial RNA was extracted us...
Embodiment 2
[0067] Example 2 Nitrogenase activity comparison of nitrogen-fixing Pseudomonas stutzeri wild-type strain, arsZ deletion mutant strain and complementing bacterial strain
[0068] 1. Construction of arsZ gene deletion mutant strain in nitrogen-fixing Pseudomonas stutzeri A1501 (P. stutzeri A1501):
[0069] First, the upstream homologous fragment of the target gene, the chloramphenicol resistance box gene, and the downstream homologous fragment of the target gene were fused into a fusion fragment with a size of about 2.1 kb by fusion PCR technology, and then the cloned fragment was subjected to BamH I and Hind III After double enzyme digestion, it was connected to the suicide vector pk18mobsacB. The constructed suicide recombinant plasmid was introduced into the wild-type A1501 bacteria through the method of triparental combination, and the suicide plasmid was integrated into the chromosome through homologous recombination with the gene on the chromosome, and the single-crossove...
Embodiment 4
[0091] Determination of half-life of nitrogenase coding gene nifD mRNA in wild-type and arsZ deletion mutants in embodiment 4
[0092] The experimental steps are as follows:
[0093] (1) Pick a single colony from the plate and inoculate it into fresh LB liquid medium containing corresponding antibiotics, culture at 30° C., 220 rpm, on a shaker overnight.
[0094] (2) Transfer the bacterial solution to fresh nitrogen-free K medium for measuring nitrogenase activity, so that the initial OD 600 is 0.3.
[0095] (3) After 6 hours of induction culture under nitrogen fixation conditions, the nitrogenase activity value was measured and recorded, and at the same time, rifampicin with a final concentration of 200 μg / ml was added to the bacterial solution to inhibit RNA synthesis. After rifampicin was treated for 0, 1, 3, 5, and 7 minutes, centrifuge rapidly at 12,000 g for 2 minutes to remove the supernatant.
[0096] (4) Add 2 times the volume of rifampicin RNA later (Sigma Company...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com