UDPG pyrophosphatase produced from aureobasidium pullulans, coding gene, carrier and application
A technology of pyrophosphatase encoding and Aureobasidium pullulans, which is applied in the fields of application, genetic engineering, and plant genetic improvement, and can solve the problems of no research results being retrieved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Cloning of embodiment 1, UDPG pyrophosphatase gene
[0032] (1) UDPG pyrophosphatase gene core fragment
[0033]Bioinformatics analysis of fungal UDPG pyrophosphatase gene sequences reported in NCBI (TrichodermareeseiQM6a, XM_006960998.1; AspergillusnigerCBS513.88, XM_001395134.2; Fusarium graminearumPH-1, XM_011317898.1; Emericellanidulans, AY850189.inado609.1; 1) After comparing the amino acid sequences using DNAMAN software, find out the conserved region and design degenerate primers. The upstream primer is: Ap190F: 5'-CCNTTYGTNGCNGAYGC-3' (SEQ ID NO.5); the downstream primer is: AP440R: 5'-NCCNGCNCCNCCRTGRTG-3' (SEQ ID NO. 6). Then take Aureobasidium pullulans NRRL12974 (source of open literature: JournalofIndustrialMicrobiology&Biotechnology.2011Sep; 38(9):1211-8.doi:10.1007 / s10295-010-0899-y.) genome as template, carry out PCR amplification, amplification condition It is: pre-denaturation at 94°C for 5 minutes; 32 cycles of denaturation at 94°C for 30 sec, annea...
Embodiment 2
[0039] Embodiment 2 constructs the Escherichia coli recombinant expression vector of UDPG pyrophosphatase
[0040] (1) Extract the total RNA of Aureobasidium pullulans NRRL12974 with the Yeast Total RNA Extraction Kit (purchased from BIOSCI Company, article number RT001), and then synthesize cDNA with the reverse transcriptase kit (purchased from BIOSCI Company, article number RT002) The first strand is used as a template for PCR amplification,
[0041] Upstream primer: 5'-ATGTCCTCCGAGATGGCAA-3' (SEQ ID NO.9);
[0042] Downstream primer: 5'-CTAGTGCTCGAGAAGTC-3' (SEQ ID NO.10).
[0043] PCR amplification conditions were: 94°C pre-denaturation for 5 min; 94°C denaturation for 50 s, 59°C annealing for 50 s, 68°C extension for 2 min, 32 cycles; 68°C extension for 5 min; 25°C for 10 min. The amplified product was connected to PMD19-TVector, transformed into E.coliJM109 competent cells, screened to obtain positive recombinants, and then sequenced to obtain a 1436bp sequence, as sh...
Embodiment 3
[0048] Expression and purification of embodiment 3UDPG pyrophosphatase
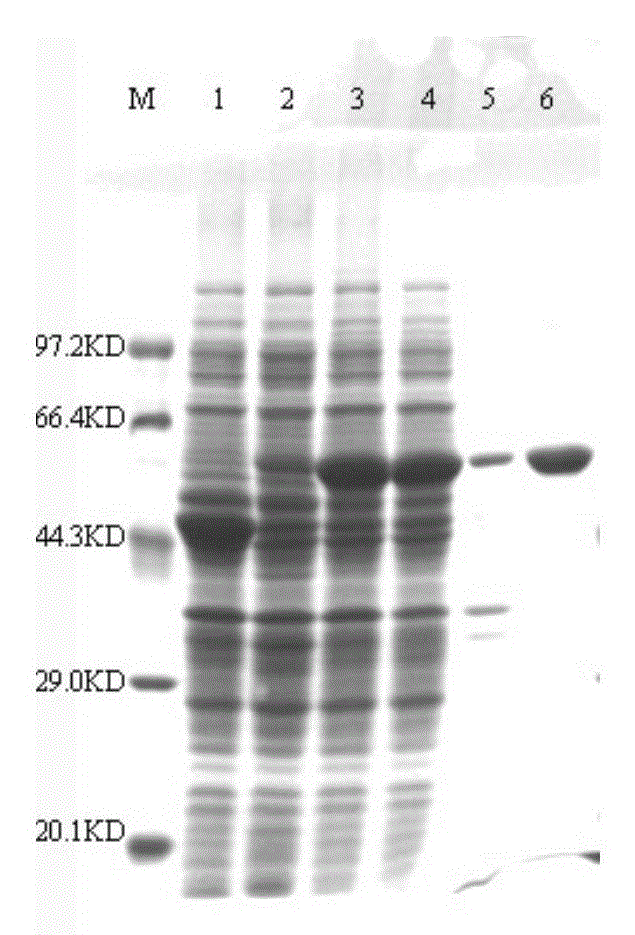
[0049] Transfer the recombinant expression vector PET28a-UGPPase constructed in Example 2 into E.coliBL21(DE3), culture the recombinant bacteria at 37°C and 180rpm until the OD600nm is about 0.6, add 0.1.mmol / L IPTG to induce After continuing to cultivate for 6 hours, collect the cells, resuspend the cells in 20mM Tris-HCl (pH7.0) buffer, lyse the cells by ultrasonication, and centrifuge to obtain the cell supernatant; pass the cell supernatant through Ni-NTA affinity chromatography (the affinity chromatography column is Ni-NTAAgarose from Qiagen Company, catnumber30210), and eluted with 150 mM imidazole aqueous solution to obtain purified protein. Then use SDS-PAGE to detect the protein, the results are as follows figure 1 shown. Depend on figure 1 It can be seen that a single band appears at about 58kDa on lane 6, which is in line with the predicted molecular weight of UDPG pyrophosphatase at 57.5K...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com