Detection method of nucleic acid
A nucleic acid and nuclease technology, which is used in biochemical equipment and methods, microbial determination/inspection, material excitation analysis, etc., to achieve the effect of improving sensitivity, simple and efficient methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
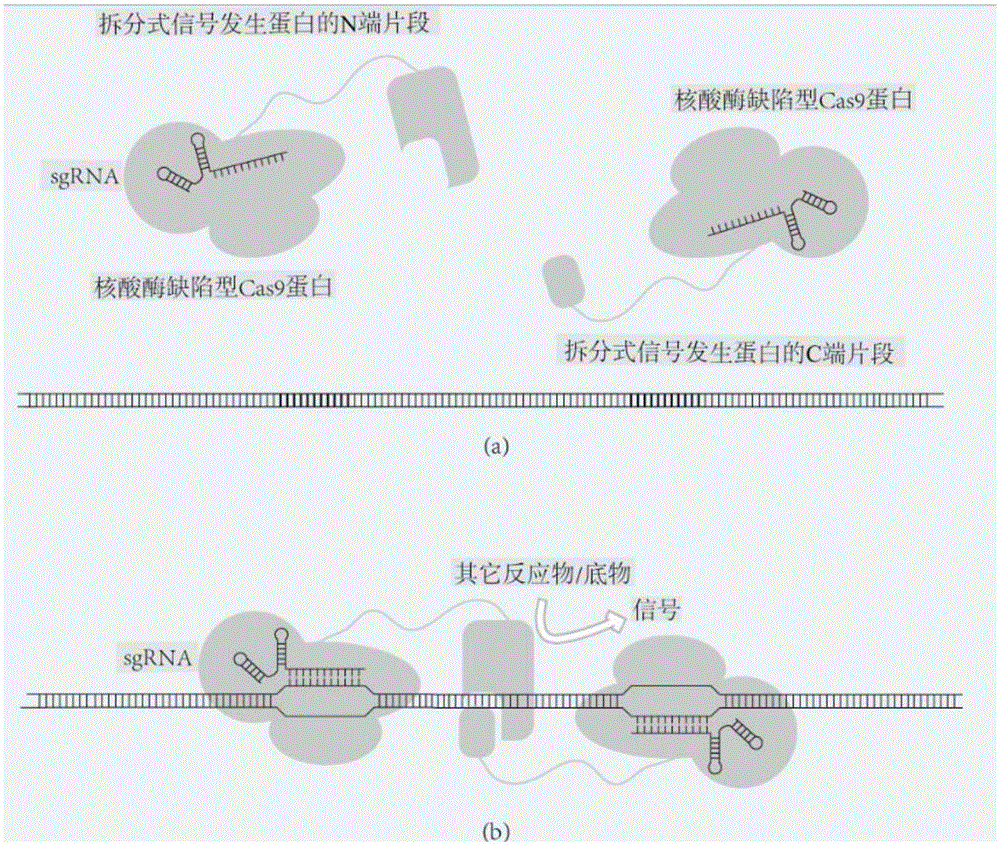
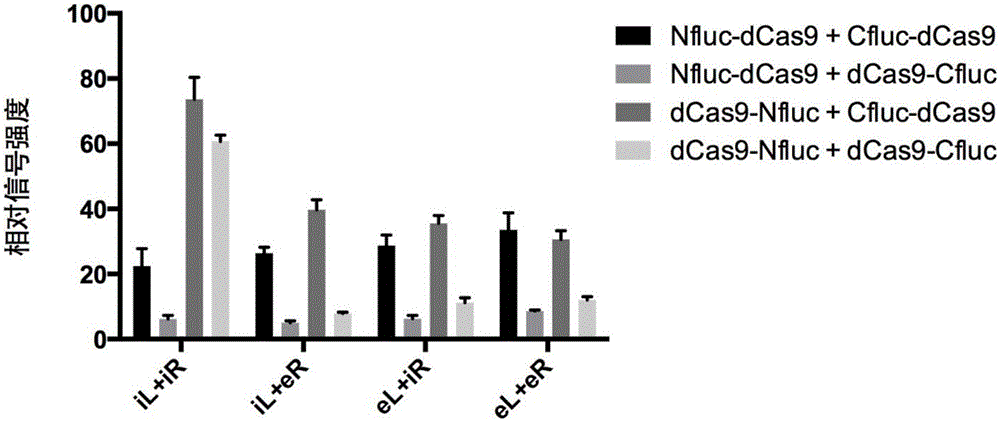
[0155] Example 1 uses a split-type firefly luciferase (nucleotide sequence shown in SEQ.ID.NO: 14) as a signal-generating protein, and its N-terminal fragment (Nfluc) has a sequence from position 1 to position 1248. The amino acid sequence encoded by the nucleotide, the C-terminal fragment (Cfluc) has the amino acid sequence encoded by the 1194th-1650th nucleotide. Use dCas9 (nucleotide sequence as shown in SEQ.ID.NO: 15) as the nuclease-deficient Cas9. The first fusion protein is a dCas9 protein (Nfluc-dCas9) that has a connecting peptide and a histidine tag and is fused with a firefly luciferase N-terminal fragment at the N-terminus, and the second fusion protein is a protein that has a connecting peptide and a histidine tag. dCas9 protein tagged and N-terminally fused to the C-terminal fragment of firefly luciferase (Cfluc-dCas9). Wherein, the nucleotide sequence encoding the connecting peptide is GGTGGCGGTGGCtctGGTGGCGGTGGCTCT (SEQ.ID.NO: 16).
[0156] The target DNA use...
Embodiment 2
[0206] In this example, the same fusion protein as in Example 1 was used, and the Bacillus subtilis genome (NCBI access NO: NC_000964) was used as the target DNA.
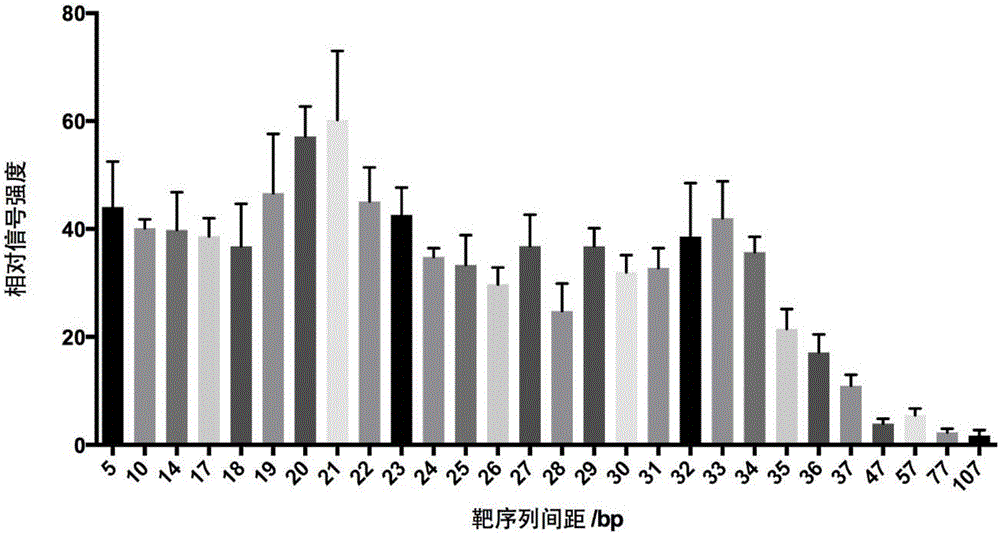
[0207] Using the same method as in Example 1, three pairs of sgRNA were designed and synthesized for the specific sequence of the Bacillus subtilis genome, named sgRNAPair6, sgRNAPair7 and sgRNAPair8, respectively. The concentration of Bacillus subtilis genomic DNA in the sample was 318.4 ng / μL. The final concentration of Bacillus subtilis genomic DNA used in the detection was 0.012 nM. A sample added with Escherichia coli genome (NCBIaccessNO: NC_000913) was used as a negative control, and the final concentration of Escherichia coli genome DNA in the detection was 0.015nM.
[0208] sgRNA pair sequence
[0209] sgRNAPair6atcatcccctcatcaatgggggcatccaccatctttgtcc
[0210] sgRNAPair7tggtatgaatcccatggttatgctgagactgttatcatat
[0211] sgRNAPair8ggaggttttggtaagggtatgtgatggtcaaggcaacaat
[0212] The results are shown ...
Embodiment 3
[0214] In this example, PCR is used as pretreatment, the same fusion protein as in Example 1 is used, and the setting of detection objects is the same as in Example 2. In the amplification negative control, the same concentration of pSB1C3 plasmid was used as the amplification template.
[0215] In the pretreatment step, conventional PCR was performed using a primer pair capable of specifically amplifying the Bacillus subtilis yraO gene fragment (forward primer: aacgaacagctgaaatggaagtgc; reverse primer: gcgcattcttggaggtgtaatatg) and the amplified product (size 2k) to recycle. The concentration of Bacillus subtilis genomic DNA in the initial sample was 318.4 ng / μL (same as Example 2). Using the recovered product as a sample (where the final concentration of DNA is 3nM), follow the same steps as in Example 2, add fusion protein and sgRNA pair, and implement the detection method of the present invention.
[0216] Such as Figure 6 As shown, compared with Example 2, using PCR a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com