Rubber tree tapping panel dryness associated protein HbMC1 and encoding gene and application thereof
A rubber tree and dead bark technology, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problem of no research report on the metacaspase family gene of rubber tree.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] Example 1. Acquisition and sequence analysis of rubber tree dead skin-related protein HbMC1 and its coding gene
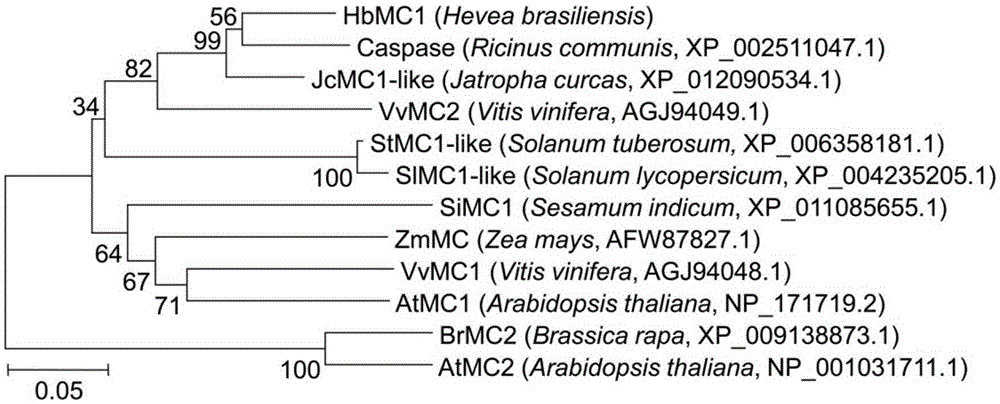
[0112] The applicant used RNA-Seq technology to analyze the difference in gene expression between healthy and dead rubber tree bark, and found that an EST was differentially expressed between healthy and dead rubber tree bark, and highly expressed in dead rubber tree bark. Then the applicant used the EST sequence as a probe to perform homologous comparison analysis on Hevea brasiliensis EST (Expressed Sequence Tags), TSA (Transcriptome Shotgun Assembly) and genome databases, and obtained a line from Hevea brasiliensis containing the complete The gene sequence of the coding region, the gene encodes the metacaspase protein, so it is named HbMC1, and the gene encoding the protein is named HbMC1 gene.
[0113] In order to verify the accuracy of the HbMC1 gene sequence, the total RNA of the bark of Hevea brasiliensis Reyan 7-33-97 was extracted using the Universa...
Embodiment 2
[0117] Example 2. Expression pattern analysis of rubber tree dead skin-related gene HbMC1
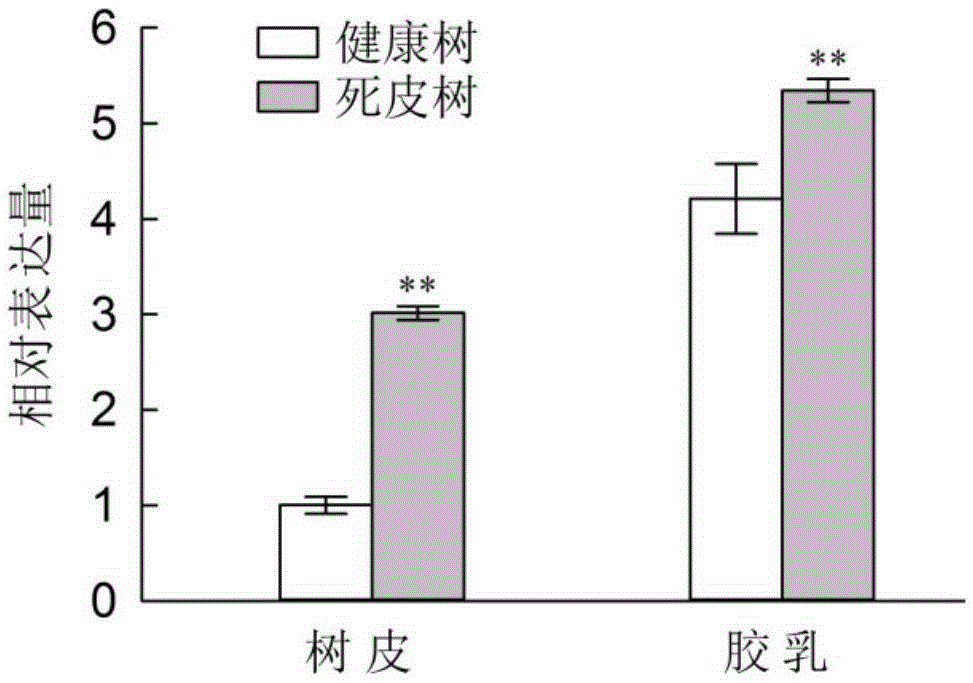
[0118] (1) HbMC1 gene is highly expressed in dead rubber tree
[0119] The applicant used the bark and latex cDNA of Hevea brasiliensis Reyan 7-33-97 healthy and dead rubber trees as templates to perform real-time fluorescent quantitative PCR. The experiment was repeated three times, and the specific steps of each repeated experiment were as follows:
[0120]Using the reverse-transcribed cDNA of total RNA extracted from the bark and latex of Hevea Reyan 7-33-97 healthy and dead bark respectively as a template, use gene-specific primers (forward primer is 5′-AGGGAGTCTCAGTTCCGGATT-3′ , the reverse primer is 5′-CGGAAGGTAGAAAAGCAGAGCA-3′) for real-time fluorescent quantitative PCR analysis. Real-time fluorescent quantitative PCR was carried out in Bio-RadCFX96 fluorescent quantitative PCR instrument, and a parallel experiment was set for 3 repetitions. reaction system: PremixExTaq TM (...
Embodiment 3
[0126] Example 3. HbMC1 gene can reduce the oxidative stress resistance of transgenic yeast and induce the death of transgenic yeast
[0127] 1. Construction of recombinant yeast
[0128] (1) Construction of yeast expression vector
[0129] A primer pair for amplifying the coding region (23rd to 1126th nucleotides at the 5' end) was designed according to sequence 2 in the sequence listing, and EcoRI and XhoI restriction sites and protective bases were introduced at the 5' ends of the forward and reverse primers, respectively. Forward primer: 5′-G GAATTC ATGCATATGCTGGTGGACTG-3' (the recognition sequence of EcoRI is underlined), reverse primer: 5'-CCG CTCGAG TCACAAAGAAAAAGGTTTTG-3' (recognition sequence of XhoI is underlined). Using the bark cDNA as a template, the target gene was amplified by PCR. The PCR reaction system is: 2 μL of cDNA template, FastPfuBuffer 5μL, 2.5mMdNTPs 2μL, forward and reverse primers (10μmol / L) each 0.5μL, FastPfuDNAPolymerase (2.5U / L) 0.5μL,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com