Melting curve analysis based oral pathogen multiple PCR (polymerase chain reaction) detection method
A melting curve analysis and detection method technology, applied in microorganism-based methods, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of low detection efficiency, long detection period, easy to cause pollution, etc. specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
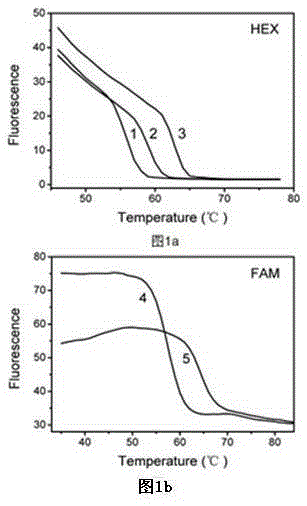
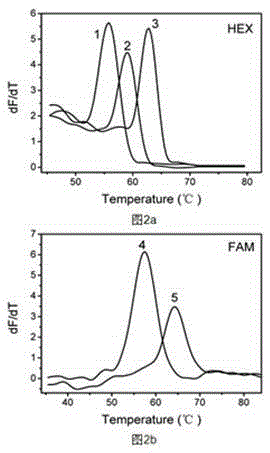
[0055] Example 1 Investigate artificial synthesis of different complementary target sequences Investigate the improved Taqman probe melting curve method to detect five oral pathogens: Porphyromonas gingivalis, Helicobacter pylori, Fusobacterium nucleatum, Streptococcus mutans, aureus staphylococcal capacity;
[0056] In this example, an improved Taqman probe for the above five kinds of oral pathogens was designed, the target sequences of the above five kinds of oral pathogens were designed, and the improved Taqman probe melting curve method was used to detect the five kinds of oral pathogens. The sequences of the modified Taqman probes and targets used were:
[0057] The probe for Porphyromonas gingivalis is 5'-HEX-catcaccacataccacagatg-BHQ2-3'
[0058] The probe for Helicobacter pylori is 5’-HEX-ctgcaacgtgacccattagcag-BHQ2-3’
[0059] The probe for Fusobacterium nucleatum is 5'-HEX-aaaatagcatactttacatattcatttt-BHQ2-3'
[0060] The probe for Streptococcus mutans is 5'-FAM-t...
Embodiment 2
[0070] Embodiment 2 investigates the artificially synthesized improved Taqman probe melting curve method after multiplex PCR to detect five kinds of oral pathogenic bacteria (Porphyromonas gingivalis, Helicobacter pylori, Fusobacterium nucleatum, Streptococcus mutans, Staphylococcus aureus at the same time )Ability.
[0071] In this embodiment, the improved Taqman probes and amplification primers for the above five oral pathogens are designed, and the whole genome DNA of the above five oral pathogens is used to investigate the improved Taqman probe melting curve method for simultaneous detection of five oral pathogens. ability of germs. The modified Taqman probes used were:
[0072] The probe for Porphyromonas gingivalis is 5'-HEX-catcaccacataccacagatg-BHQ2-3'
[0073] The probe for Helicobacter pylori is 5’-HEX-ctgcaacgtgacccattagcag-BHQ2-3’
[0074] The probe for Fusobacterium nucleatum is 5'-HEX-aaaatagcatactttacatattcatttt-BHQ2-3'
[0075] The probe for Streptococcus m...
Embodiment 3
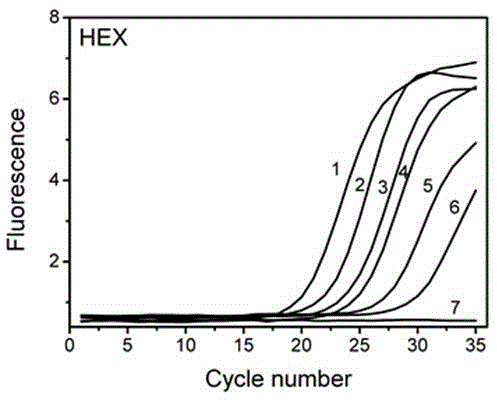
[0094] Example 3 investigates the sensitivity of the artificially synthesized and improved Taqman probe melting curve method to detect different concentrations of Fusobacterium nucleatum plasmid standard substances after multiplex PCR.
[0095] In this example, the improved Taqman probes and amplification primers for the above five oral pathogenic bacteria were designed, and the plasmid standards for the complementary target sequence of Fusobacterium nucleatum were artificially synthesized, and the melting curve of the improved Taqman probe was investigated to detect the complementation of Fusobacterium nucleatum. Detection sensitivity of the target sequence.
[0096] The modified Taqman probes used were:
[0097] The probe for Porphyromonas gingivalis is 5'-HEX-catcaccacataccacagatg-BHQ2-3'
[0098] The probe for Helicobacter pylori is 5’-HEX-ctgcaacgtgacccattagcag-BHQ2-3’
[0099] The probe for Fusobacterium nucleatum is 5'-HEX-aaaatagcatactttacatattcatttt-BHQ2-3'
[0100...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Upstream primer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com