Prawn covert mortality nodavirus fluorescent quantitative RT-PCR detection method
A Nodamura virus and fluorescence quantitative technology, which is applied in the field of fluorescence quantitative RT-PCR detection of prawn stealing Nodamura virus. , complex operations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1 RNA standard product construction
[0065] 1) Using the CMNV-positive disease RNA as a template, reverse transcription PCR was performed using primers 1 and 2 to obtain a PCR product with a fragment size of 179 bp, and the PCR product was gel-recovered and purified.
[0066] 2) T-VectorpMD TM 20 was used as the transcription vector of the purified PCR product, and then transformed into DH5α competent cells.
[0067] 3) Extract the plasmid using a plasmid extraction kit (Biomed).
[0068] 4) Using EcoRI (TaKaRa Company) for 2 hours at 37° C. to linearize the plasmid. The system is:
[0069]
[0070] 5) Use SP6RNAPolymerase (TaKaRa Company) for in vitro transcription, react at 37°C for 1 hour, the system is:
[0071]
[0072] 6) The synthesized RNA was degraded with DNaseI for plasmid DNA, and reacted at 37°C for 30 minutes. The system was as follows:
[0073]
[0074] 7) Carry out phenol chloroform extraction and purify the extracted RNA
[00...
Embodiment 2
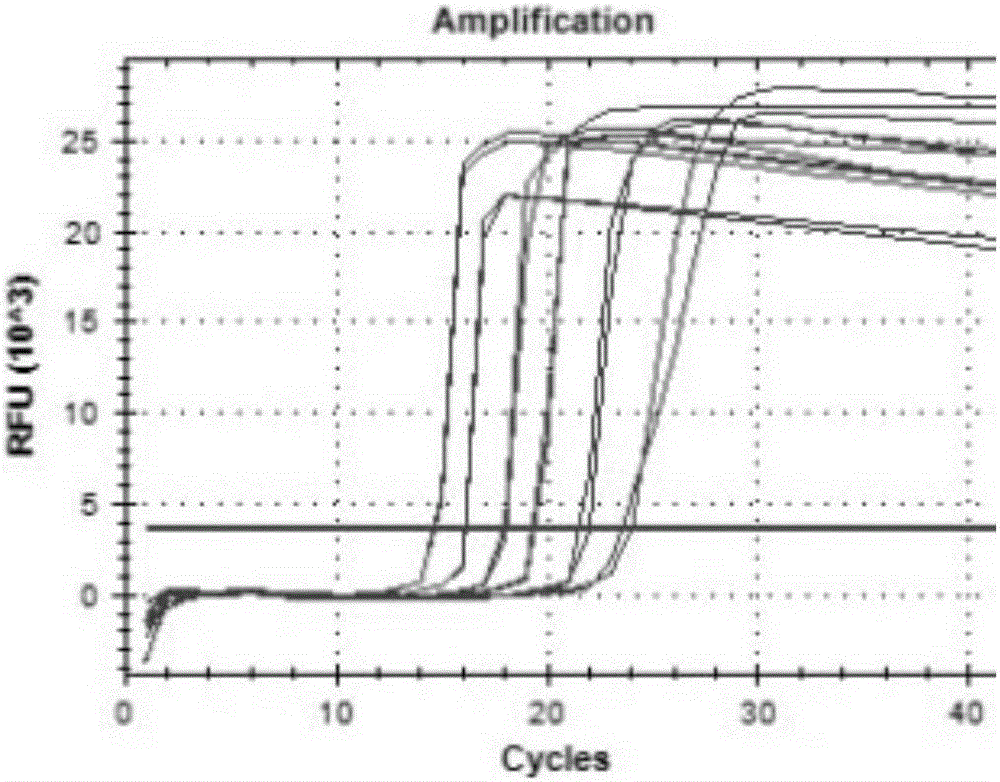
[0085] The making of embodiment 2 standard curve
[0086] Using the concentration of synthetic RNA measured in Example 1, the copy number of synthetic RNA was calculated. Use RNaseFree water to carry out 10-fold gradient dilution of RNA, dilute to 10 7 ~10 2 copies / μL, used as a standard to make a standard curve.
[0087] One-step fluorescent quantitative RT-PCR kit was used, and the reaction solution was prepared according to the instructions of the kit. The total volume of RT-PCR reaction solution was 25 μL: reaction buffer 12.5 μL, TaqHS mixed solution 1 μL, PrimeScript PLUS RTase mixed solution 1 μL, ROX reference The dye is 1ul, the probe is 0.2-0.5μL (10mmol / L), and the two primers and RNA are added at the same time. The final concentration of the primer is 1μL (10mmol / L), and the RNA is 1μL. Use RNase-free water as a supplement 25 μL.
[0088] Use the Roche LightCycler480II real-time fluorescence quantitative PCR system to perform RT-PCR reverse transcription reacti...
Embodiment 3
[0089] Embodiment 3 clinical verification of shrimp samples
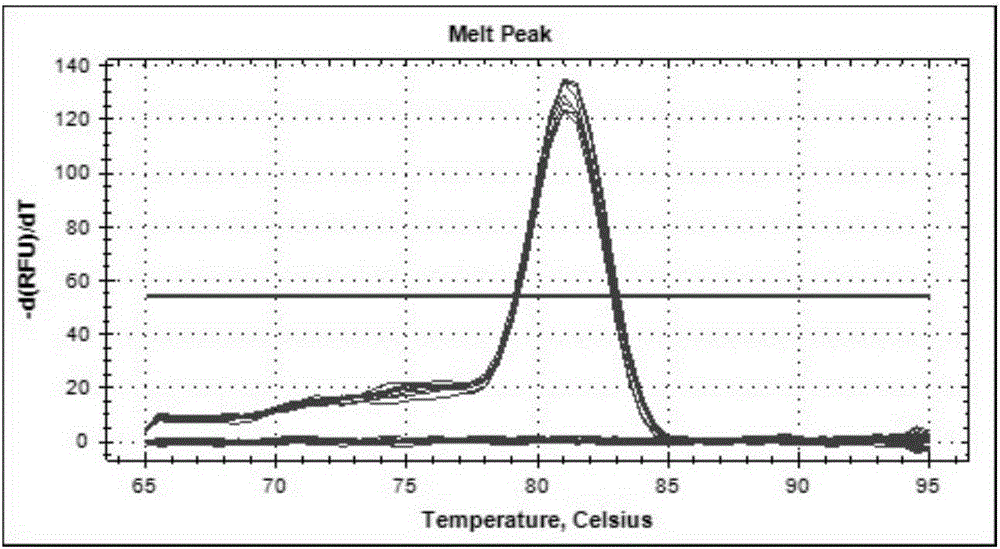
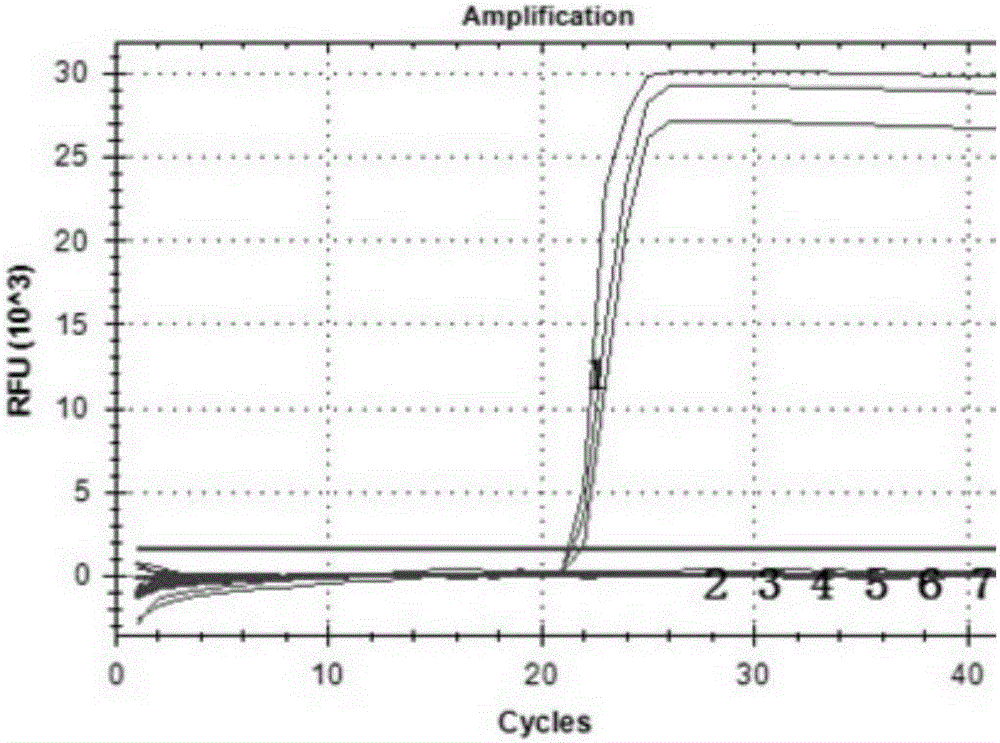
[0090] For 15 shrimp samples collected from Shandong, Hebei, Tianjin, Zhejiang and Hainan in 2013-2014, RNA was extracted using a commercial RNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.). The reaction premix was prepared according to the above method, and the RT-PCR reaction was carried out according to the preferred amplification conditions. The results showed that 9 samples were amplified, and the Ct values of the amplification curves were all less than 35 and the Tm values were 81.53±0.14°C, so the positive rate of confirmed samples was 60%. Nested reverse transcription PCR verification was carried out on these 15 samples, and the results showed that 8 of the above 15 samples had positive bands, and the positive rate was 53%. Therefore, the fluorescent quantitative RT-PCR method of the present invention has higher sensitivity than the common nested reverse transcription PCR method. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com