Single-molecule analysis method for detecting methylated DNA
An analytical method and methylation technology, applied in the direction of electrochemical variables of materials, etc., can solve the problems of complexity, high technical requirements, poor reproducibility of results, etc., and achieve the effect of small sample volume, high sensitivity, and simple and easy operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Specifically, the detection of methylated hairpin DNA by the present invention comprises the following steps:
[0047] (1) Install the pre-punched Telfon membrane in the center of the electrolytic cell, divide the electrolytic cell into two parts, and drop a mixture of pentane and hexadecane containing 1% hexadecane around the hole with a capillary tube. Add 4M tetramethylammonium chloride, 10mM Tris-HCl, pH8.5 buffer solution into the tank, add a small amount of phospholipid dropwise above the liquid surface, add buffer solution to make the liquid surface cross the small hole, and form a stable phospholipid bilayer. The electrolytic cell is cis-added with wild-type α-hemolysin, which is inserted into the phospholipid bilayer to form a nano single channel.
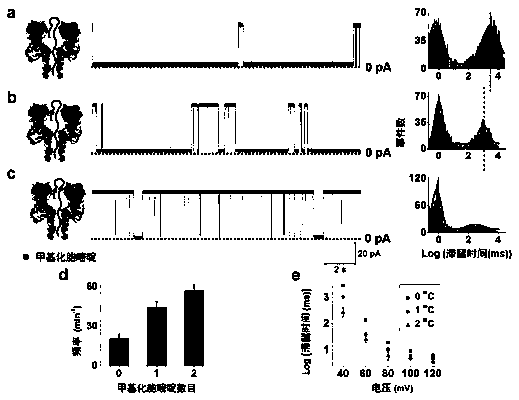
[0048] (2) Add the hairpin DNA containing 0 / 1 / 2 methylated cytosine in the stem region in cis to the electrolytic cell, stir evenly, and detect the DNA current signal under positive voltage (+40mV~+120mV). The tail...
Embodiment 2
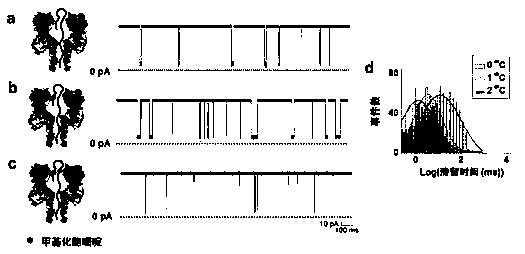
[0053] Potassium chloride is a traditional nanopore single-molecule detection electrolyte, similar to Example 1, under the condition that other experimental conditions are kept constant, the detection results of methylated hairpin DNA in potassium chloride electrolyte are as follows figure 2 as shown, figure 2 -a, b, c are schematic diagrams of DNA detection principle (left), single-channel current signal diagram (right). figure 2 -a is hairpin DNA that does not contain methylated cytosine; figure 2 -b is hairpin DNA containing 1 methylated cytosine; figure 2 -c is hairpin DNA containing 2 methylated cytosines; figure 2 -d is the distribution statistical graph of the residence time of the three kinds of hairpin DNA.
[0054] First of all, in the KCl electrolyte, the difference in the retention time of the three hairpin DNAs is only ~10ms, which is much smaller than the ~2600ms in the tetramethylammonium chloride electrolyte; and, the retention time of the hairpin DNA ...
Embodiment 3
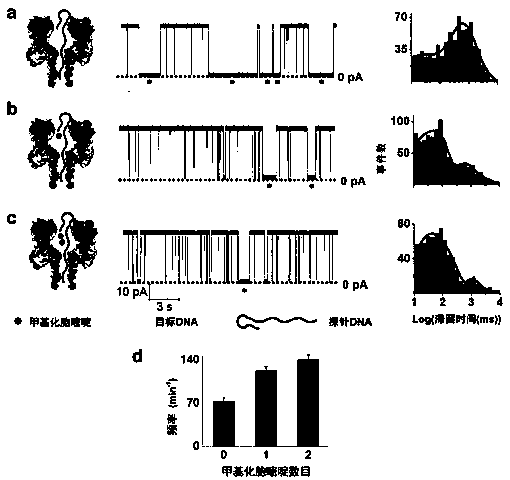
[0056] Detection of methylated single-stranded DNA includes the following steps:
[0057] (1) Hairpin probe DNA and target DNA were mixed at a concentration ratio of 1:1, heated at 95°C for 5 minutes, and cooled naturally.
[0058] (2) Install the pre-punched Telfon membrane in the center of the electrolytic cell, divide the electrolytic cell into two parts, and drop a mixture of pentane and hexadecane containing 1% hexadecane around the hole with a capillary tube. Add 4M Tetramethylammonium Chloride, 10mM Tris-HCl, pH8.5 buffer solution into the tank, add a small amount of phospholipid dropwise above the liquid surface, add buffer solution to make the liquid surface submerge the small holes, and form a stable phospholipid bilayer. The electrolytic cell is cis-added with wild-type α-hemolysin, which is inserted into the phospholipid bilayer to form a nano single channel.
[0059] (3) Add the mixture of hairpin probe DNA and target DNA containing 0 / 1 / 2 methylated cytosine in c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com