Engineering plants for efficient uptake and utilization of urea to improve crop production
A technology of urea and plants, applied in the field of molecular biology, can solve problems such as low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0204] Example 1: Possible urea from maize and from different microbial and lower plant sources Identification and evolution analysis of transporters
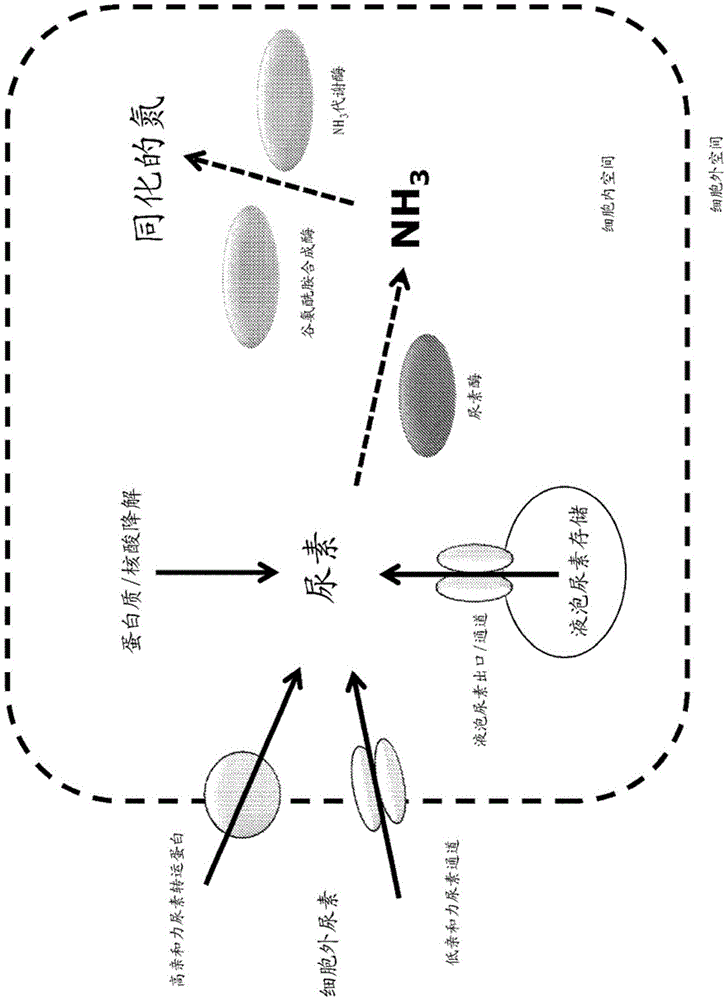
[0205] Several classes of proteins with the ability to translocate urea throughout biofilms have been identified in various species ranging from bacteria to higher eukaryotes such as plants. Proteins encoding eukaryotic DUR3 high-affinity urea transporters (SEQ ID NOs: 1-187), prokaryotic UREI urea channels (SEQ ID NOs: 188-193), and aquaporin-like NIP / TIP / PIP proteins (SEQ ID NOs: 194-223) were identified Multiple genes of possible homologues. Sequences of possible DUR3 homologues were obtained from a variety of sources (mainly focusing on microorganisms, fungi, and lower photosynthetic plants), and evolutionary analyzes were performed to examine the relationship between transporters ( image 3 ). Potential maize Dur3 homologues were also identified based on sequence homology to urea transporters such as the Arabi...
example 2
[0213] Example 2: Identification of possible urease genes from various plant and microbial sources
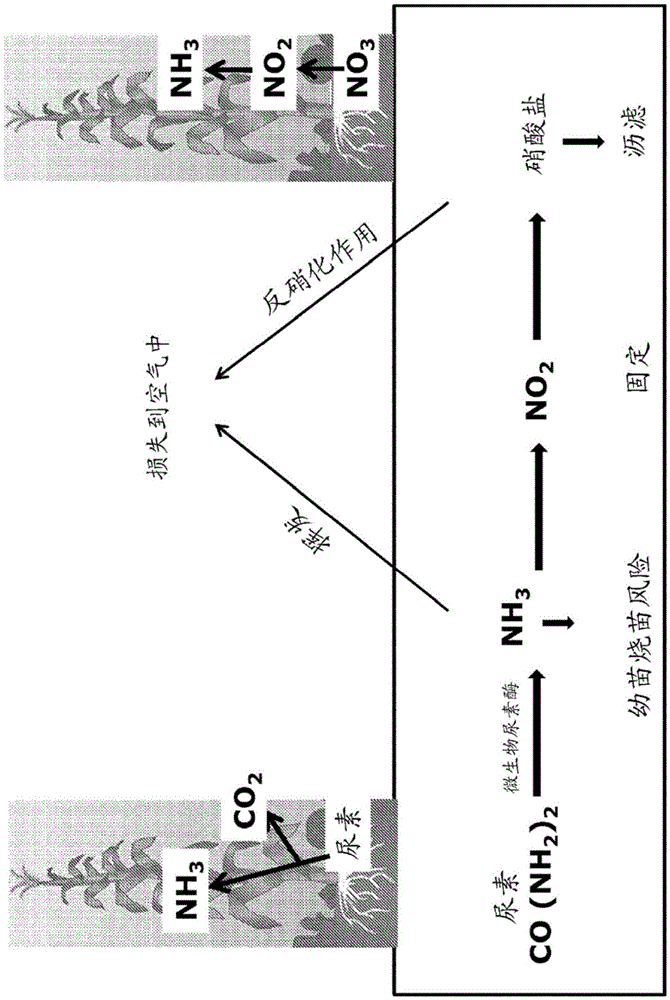
[0214] Urease is an enzyme necessary for the breakdown of urea into ammonia that can be utilized by cells. Multiple genes encoding possible urease proteins (SEQ ID NO: 251-298) were identified by bioinformatics methods. The analysis mainly focused on a single subunit urease protein encoded by only one transcript, and the sequences of 48 known or putative ureases were obtained from various sources, mainly fungal or plant sources. Evolutionary analysis was performed to detect the relationship between known and putative urease proteins ( Figure 4 ). While expression of the urease protein alone is expected to be sufficient to enhance urease activity in plants, co-expression of urease with its corresponding urease accessory protein can enhance this enzymatic activity. In bacteria, these proteins are easy to identify because they fall within the same operon as the urease ...
example 3
[0215] Example 3: Identification of possible GS genes from maize and other sources
[0216] Once urea is broken down into its constituent parts by urease, the released ammonia is assimilated into amino acids mainly through the action of glutamine synthetase. Multiple genes encoding possible glutamine synthetase proteins have been previously identified (see US Patent Application No. 12 / 607,089 published May 6, 2010). The sequences of two known glutamine synthetases can also be used: the GS1-3 subtype from maize and from Arabidopsis. Other GS1, GS2 or GS3-type glutamine synthetase proteins from plants or other sources can also be used to form transgenic plants with enhanced ability to utilize urea as a nitrogen source.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com