Method for raising S-adenosyl-L-methionine production level by saccharomyces cerevisiae metabolic engineering
A technology of metabolic engineering transformation and adenosylmethionine, which is applied in microorganism-based methods, biochemical equipment and methods, fermentation, etc., can solve problems such as insufficiency, and achieve improved knockout efficiency, good technical effects, and excellent food safety. The effect of biological properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
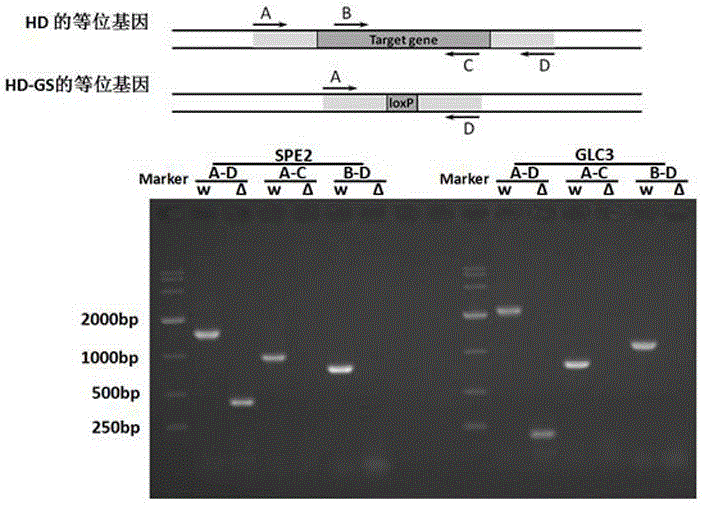
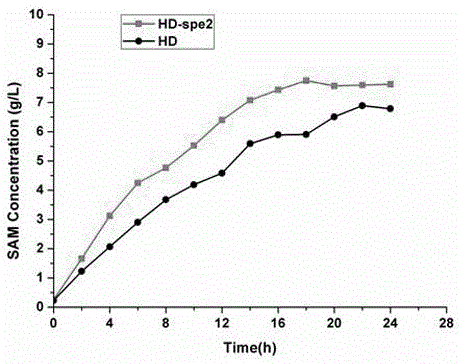
[0049] Example 1: Taking diploid Saccharomyces cerevisiae strain HD as an example, construct homozygous knockout SPE2 gene and compare SAM yield with HD.
[0050] 1. PCR construction of knockout box. The method is as follows: using the plasmid pUG6 as a template, using primers to perform PCR reactions on SPE2-up and SPE2-down, the PCR products are purified and recovered by a DNA purification kit, confirmed by DNA electrophoresis, and finally used as a knockout frame for SPE2 gene knockout.
[0051] 2. LiAC conversion. The method is as follows: Pick a single colony of the original strain HD and place it in a 20mLYPD shake flask, rotate at 200rpm, cultivate overnight at 30°C, then transfer 2mL to a 50mLYPD shake flask and continue to cultivate for 3-4 hours, OD 600 Centrifuge to collect the bacteria at about 1. Wash once with 20mL sterile water, centrifuge, and remove the supernatant; then wash once with 0.1M LiAC, centrifuge, and remove the supernatant; resuspend the bacteria with ...
Embodiment 2
[0059] Example 2: Knock out the SPE2 gene in the model strain BY4741 and verify its yield.
[0060] Methods as below:
[0061] 1. PCR construction of knockout box. The method is as follows: using the plasmid pUG6 as a template, using primers to perform PCR reactions on SPE2-up and SPE2-down, the PCR products are purified and recovered by a DNA purification kit, confirmed by DNA electrophoresis, and finally used as a knockout frame for SPE2 gene knockout.
[0062] 2. LiAC transformation and PCR verification. The method is as follows: Pick a single colony of the original strain BY4741 and place it in a 20mLYPD shake flask, rotate at 200rpm, cultivate overnight at 30°C, then transfer 2mL to a 50mLYPD shake flask and continue to cultivate for 7-8 hours, OD 600 Centrifuge to collect the bacteria at about 1. Wash once with 20mL sterile water, centrifuge, and remove the supernatant; then wash once with 0.1M LiAC, centrifuge, and remove the supernatant; resuspend the bacteria with 1mL0.1ML...
Embodiment 3
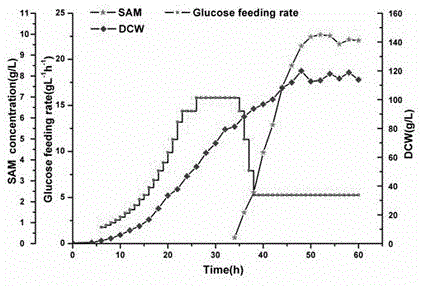
[0066] Example 3, taking the diploid Saccharomyces cerevisiae strain HD as an example, a strain with both GLC3 and SPE2 knocked out was constructed, and SAM yields were compared.
[0067] 1. Similar to the first step of the example, first knock out the GLC3 gene: use pUG6 as a template PCR to construct a GLC3 knockout box. After LiAC transformation, G418 plate screening and PCR verification, the GLC3 knockout heterozygotes are obtained, and then the heterozygotes are cultured for sporulation , Isolate the spores, screen on the G418 plate to obtain the homozygous GLC3 knockout, and perform PCR verification and confirmation.
[0068] 2. Recover the marker gene. The method is as follows: place the homozygous single colony knocked out of GLC3 in a 20mLYPD shake flask, rotate at 200 rpm, cultivate overnight at 30°C, then transfer to a 2mL 50mLYPD shake flask and continue to incubate for 3-4 hours, when the OD600 is about 1. Collect the bacteria by centrifugation. Wash once with 20mL s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com