Method for production of phenol by heterogenous metabolism path and application thereof
A metabolic pathway, phenol technology, applied in the field of clean production of phenol, can solve problems such as serious environmental pollution and non-renewability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
[0027] Construction of Metabolic Pathway for Microbial Heterologous Synthesis of Phenol
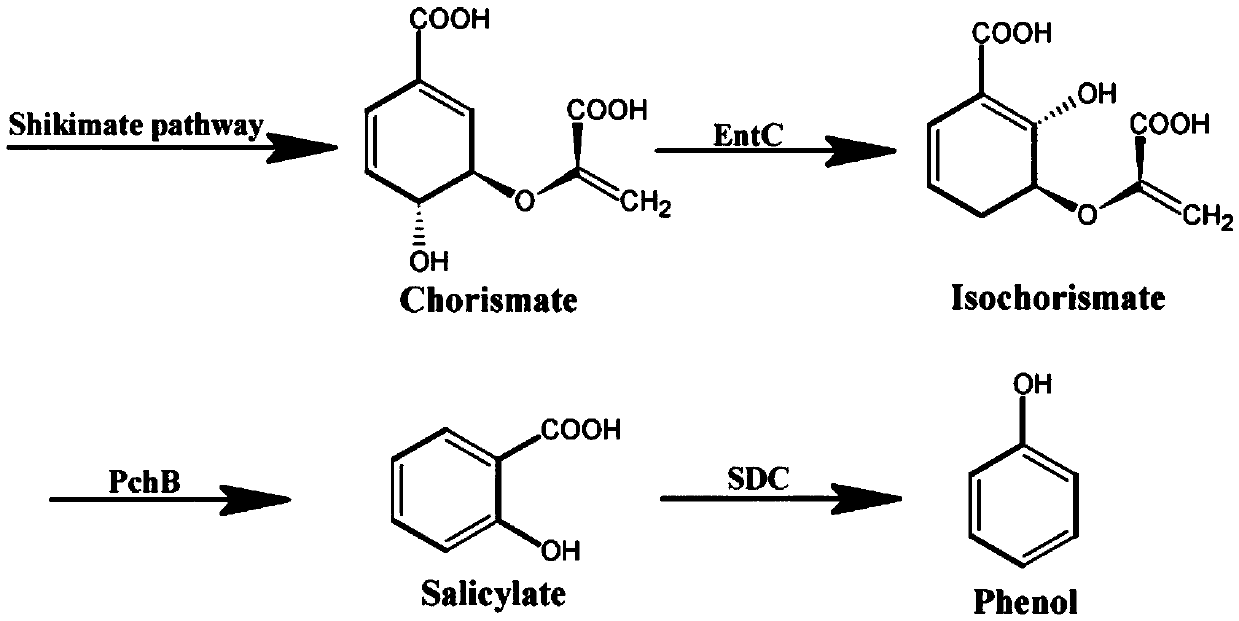
[0028] Aromatic compounds are naturally synthesized by many bacteria. The pathway for the synthesis of aromatic amino acids is called the shikimate pathway. In the shikimate pathway, glucose undergoes a central metabolic pathway to synthesize phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P). These two compounds are enzymatically formed into 3-deoxy-D-arabinosyl-7-phosphate (DAHP), which after several steps of reaction forms chorismate. Starting from chorismate, the pathway branches into several branches to form a variety of aromatic compounds, including phenylalanine, tyrosine, tryptophan, folic acid, etc. So far, the shikimate pathway has been used to synthesize a variety of aromatic compounds, some of which have been commercialized. The seven enzymes of the shikimate pathway were originally discovered through studies on bacteria, mainly Escherichia coli and Salmonella typhim...
specific Embodiment 2
[0030] Introduction of heterologous metabolic pathways into phenol-producing microorganisms
[0031] The genomes of Escherichia coli, Pseudomonas fluorescens, and Klebsiella pneumoniae were extracted respectively, and the target genes, entC, pchB, and kpBDC were obtained by PCR from the genomes using the designed primers, as shown in Table 2. Then use the corresponding enzymes to digest the fragments and vectors, and perform gel recovery or column recovery on the digested fragments, and then insert the target gene into the plasmid pZE12-luc (high copy) and plasmid pCS27 (medium copy) to obtain The connecting fluid is spare.
[0032] Preparation of competent cells by electroporation: Inoculate single colonies of Escherichia coli BW25113 and QH4 into 3 mL of liquid LB medium, culture overnight at 37° C. and 250 rpm. Then inoculate 500 μL of bacterial solution into 50 mL of LB medium, culture at 37° C. with shaking at 250 rpm until OD=0.4-0.6. Centrifuge the bacterial solution ...
specific Embodiment 3
[0039] Fermentation of phenol-producing microorganisms
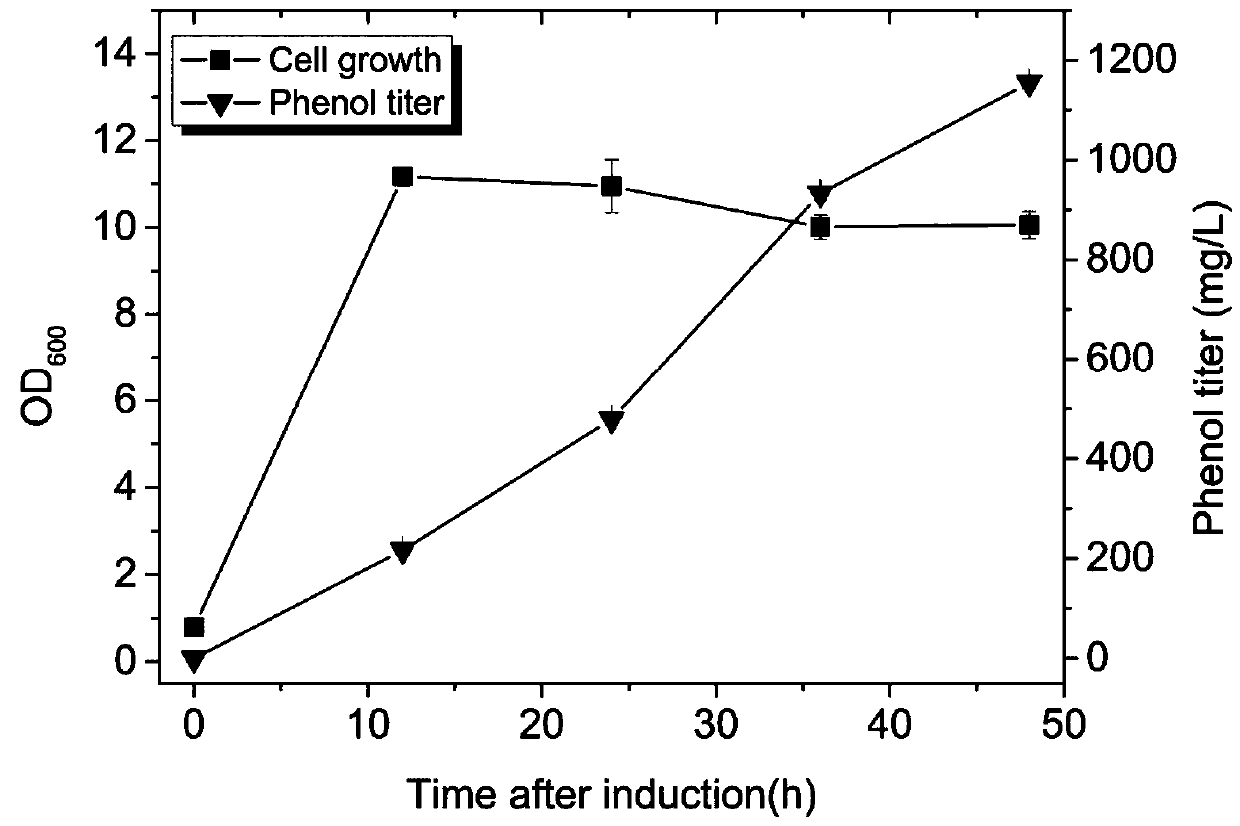
[0040] Pick a single colony from the plate in the genetic engineering of Escherichia coli, connect it to 3mL liquid LB, add ampicillin and kana antibiotics to the medium, the final concentration of antibiotics is 100mg / L, shake culture at 37°C, 250rpm, and then Transfer the bacterial liquid to 50mL liquid M9Y medium, add ampicillin and kana antibiotics to the medium, the final concentration of antibiotics is 100mg / L, shake the culture until OD600 reaches 0.6, add IPTG, IPTG to the medium The final concentration in the medium was 0.25 mM, induced at 30°C. After induction, samples were taken every 12hrs to measure OD, and the concentrations of phenol and salicylic acid were determined by high performance liquid chromatography.
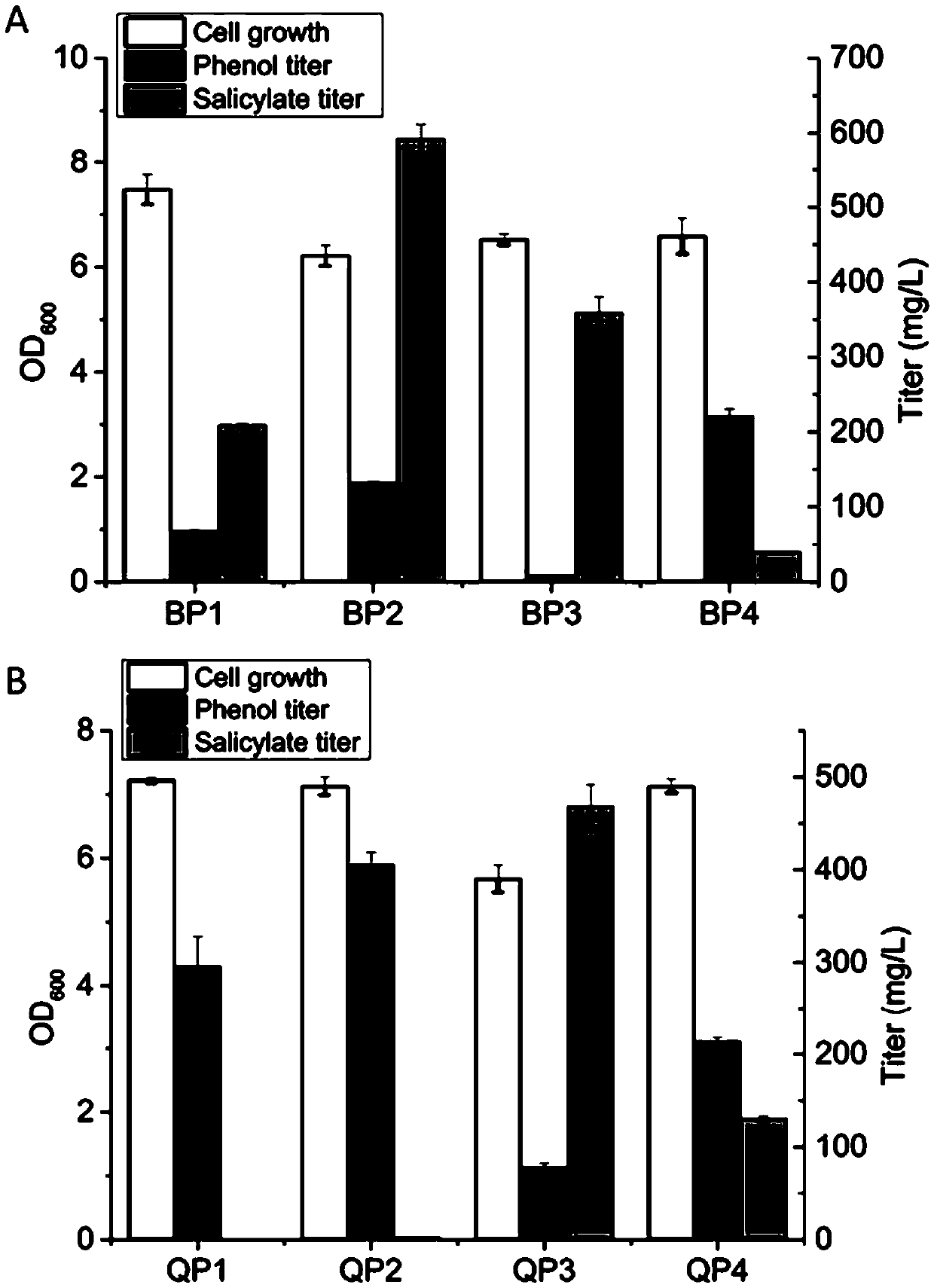
[0041] The fermentation results showed that the plasmid pZE-EP-BDC was introduced into Escherichia coli BW25113, and the strain was defined as BP1. The BP1 strain could only produce 67.65±1.65mg / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com