A method for detecting the position of large segment repeats in Saccharomyces cerevisiae chromosome
A technology for Saccharomyces cerevisiae and Saccharomyces cerevisiae strains is applied in the field of detecting the repetitive position of large segments of Saccharomyces cerevisiae chromosomes, which can solve the problems of cumbersome experimental data analysis work, and achieve the effects of short experimental period, simple operation and rapid detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] 1. Culture medium preparation
[0050] The formula of SC-Lys-Met auxotrophic culture plate is synthetic yeast nitrogen source YNB 6.7g / L, glucose 20g / L, default mixed amino acid powder of lysine and methionine 2g / L, agar powder 20g / L.
[0051] The formula of YPD liquid medium is yeast extract 10g / L, peptone 20g / L, glucose 20g / L.
[0052] The formula of SC+5-FOA medium is synthetic yeast nitrogen source YNB 6.7g / L, glucose 20g / L, mixed amino acid powder 2g / L, 5-fluoroorotic acid 1g / L, agar powder 20g / L.
[0053] 2. Experimental method
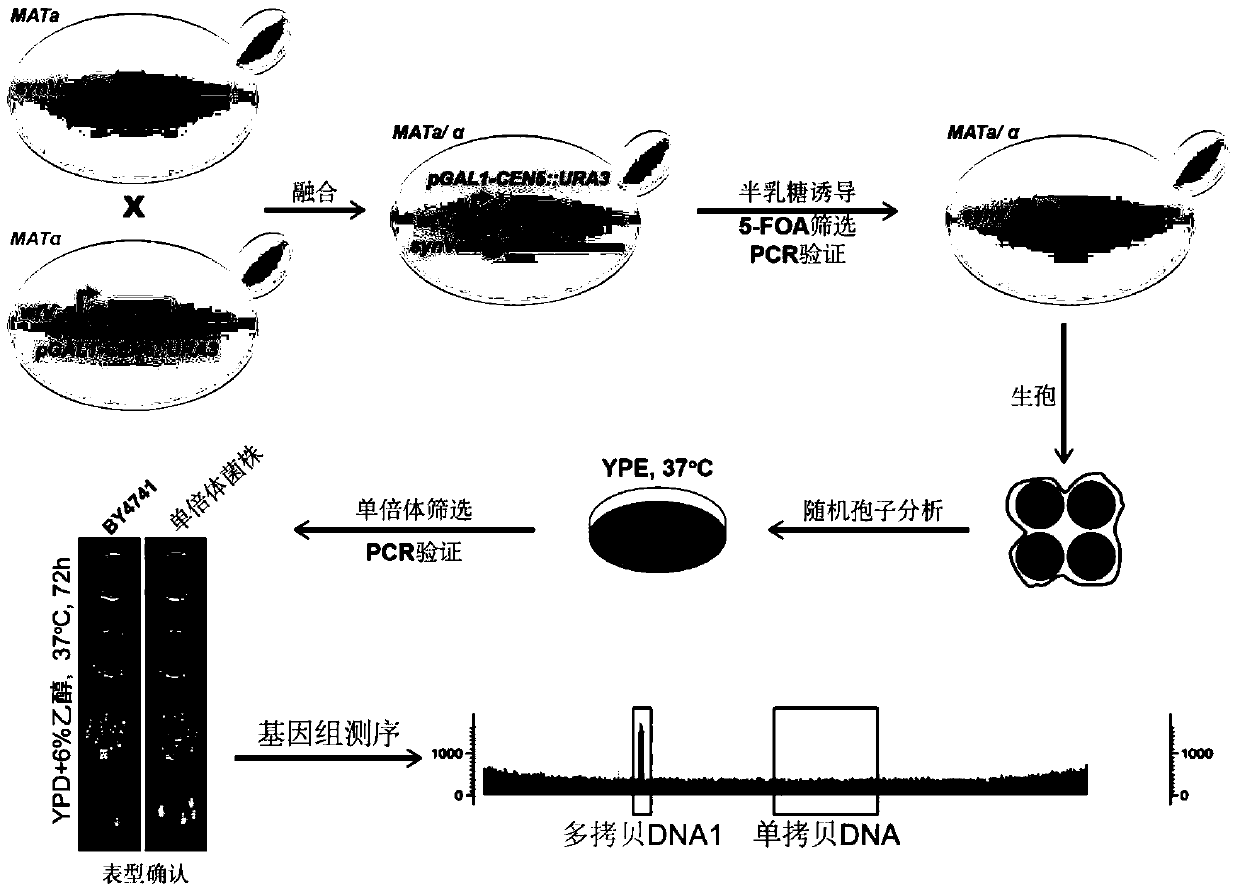
[0054] 1. Sequence the entire genome of the tested Saccharomyces cerevisiae, and the results are as follows: figure 1 , showing that there is a small repeating region (multi-copy DNA1) and two large repeating regions (multi-copy DNA2) on chromosome V, and the chromosome is named "source chromosome V (synV)".
[0055] 2. Using the method reported in literature (Reid, R.J., et al.Genetics (2008), 180, 1799-1808), add the GAL1 promoter an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com