CYP2C9*3 detection parting kit based on probe AllGlo and parting method of CYP2C9*3 detection parting kit
A kit and probe technology, applied in the field of single nucleotide polymorphism, can solve the problems of differences in effective treatment doses, reduce enzyme activity, etc., and achieve the effects of low detection price, improved Tm value, and good quenching effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
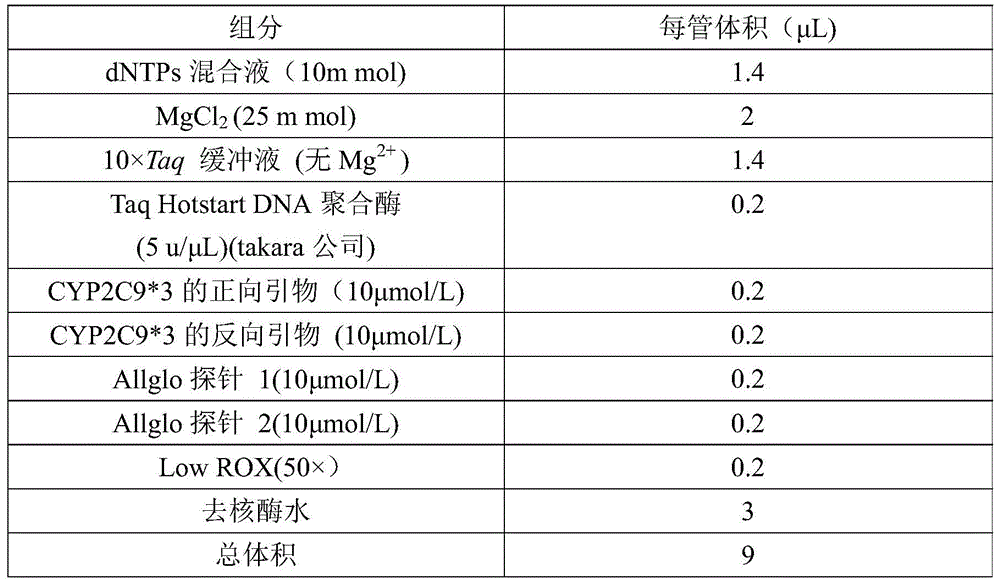
[0038] The CYP2C9*3 detection and typing kit based on the AllGlo probe of the present invention comprises the following components: real-time fluorescence quantitative PCR reagent, positive control and negative control.
[0039] ①Reagents for real-time fluorescent quantitative PCR include the following components: 10×Taq buffer (10×Taq buffer includes 100mmol Tris-HCl and 500mmol KCl), 10mmol MgCl 2 , 5u / μL TaqHotstart DNA polymerase, 10mmoldNTPs mixture, 50×LowROX, 100mL nuclease-free water, 10μmol / LCYP2C9*3 specific forward primer, 10μmol / LCYP2C9*3 specific reverse primer and AllGlo probe.
[0040] ② Positive controls include: positive homozygous control substance 1, positive homozygous control substance 2, and positive heterozygous control substance. The positive homozygous control substance 1 is a DNA sample of CYP2C9*3 type AA, the positive homozygous control substance 2 is a DNA sample of CYP2C9*3 type CC, and the positive heterozygous control substance is CYP2C9* 3 DNA...
Embodiment 2
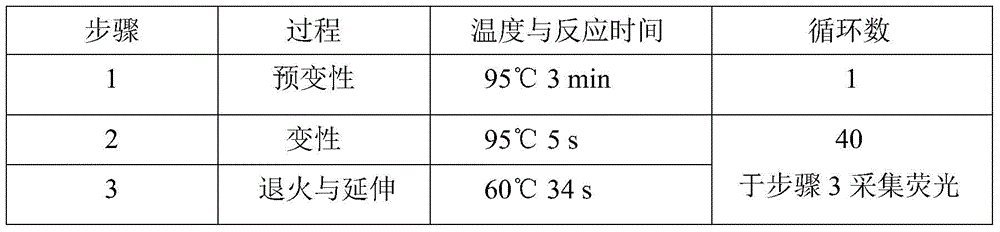
[0043] The detection and typing of CYP2C9*3 in peripheral blood includes the following steps:
[0044] 1. Collect EDTA anticoagulated peripheral blood:
[0045] Take 2 mL of fresh blood samples and put them into EDTA anticoagulant tubes and divide them into multiple tubes. Each tube is divided into 400-500 μL, and 200 μL is used for DNA extraction. The rest of the whole blood samples are stored at -80°C.
[0046] 2. Extract DNA:
[0047] Use Genomic Blood DNA Extraction Kit (DP348) produced by Tiangen Biotechnology Co., Ltd. to extract DNA from the sample (whole blood) according to the operating instructions, 200μL of LEDTA anticoagulated whole blood according to the instructions, and finally use 50μL of elution buffer TB dissolves the DNA, and measures the concentration and purity on the nucleic acid spectrophotometer NanoDrop2000. The ratio of purity A260 / A280 is between 1.7 and 1.9, and the extracted DNA with a concentration of 10-50 ng / μL is used for SNP typing detection....
Embodiment 3
[0062] Example 3. SNP detection and typing of tissue samples
[0063] 1. Tissue sample processing:
[0064] Tissues (the amount of spleen tissue should be less than 10 mg) should be crushed and processed into a cell suspension, then centrifuged at 10,000 rpm (~11,200×g) for 1 min, and the supernatant was poured out, and 200 μL of buffer GA (Tiangen DNA Extraction Kit DP304) was added, and shaken until completely suspended;
[0065] 2. DNA extraction and enrichment of tissue samples:
[0066] Use the DNA extraction kit (DP304) produced by Tiangen Biotechnology Co., Ltd. to extract DNA from tissue samples according to the operating instructions, and finally dissolve the DNA with 50 μL of elution buffer TE, and measure the concentration on the nucleic acid spectrophotometer NanoDrop2000 and purity;
[0067] 3. SNP detection and typing of tissue samples.
[0068] For subsequent steps, refer to steps 3-4 of Example 2.
[0069] Applicable range:
[0070] The AllGlo probe-based...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com