Oryza sativa histone methyltransferase as well as encoding gene and application thereof
A technology of methyltransferase and histone, which is applied in the fields of transferase, application, genetic engineering, etc., can solve the problems of research lag and few, and achieve the effects of delayed flowering time, short growth and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1. Rice histone methyltransferase gene SDG701 the acquisition
[0027] The SDG2 gene sequence of Arabidopsis thaliana published in GenBank (GenBank number: NC_003075.7) was compared with the homologous sequence, and the homologous sequence was obtained from the rice genome, and according to the 5' and 3' end sequences of the homologous sequence A pair of primers were designed, and the primer sequences were: 5'-ATGCCGGACAAGGGGGAGA-3' (SEQ ID No: 3) and 5'-CTAGCCTAGGAAGACGTTGGATCTTG-3' (SEQ ID No: 4).
[0028] The total RNA of etiolated rice seedlings was extracted (Promega, SVtotalRNAisolationsystem), and the total RNA of rice was used as a template to synthesize cDNA with AMV reverse transcriptase (TaKaRa) (according to the user manual of PlantRT-PCRKit2.01 (TaKaRa)). PCR Amplification of Rice Histone Methyltransferase Gene Using cDNA as Template SDG701 full-length cDNA sequence. The 50 μl PCR reaction system contains: 2 μl template, 1 μl high-fidelity enzy...
Embodiment 2
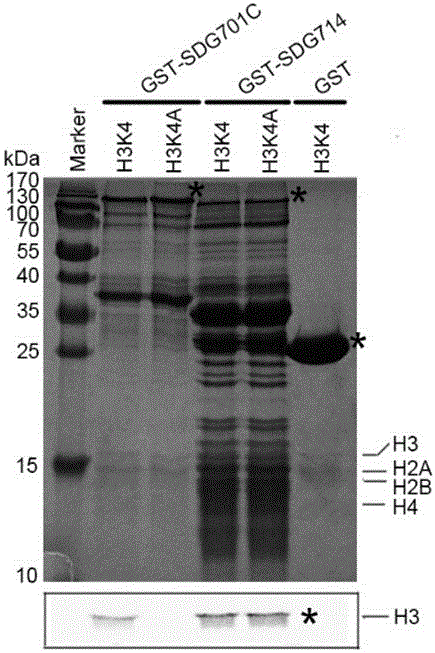
[0029] Example 2. Rice histone methyltransferase gene SDG701 protein activity assay
[0030] Using the pUC19-SDG701 obtained in Example 1 as a template, PCR amplification SDG701From the nucleotide sequence at position 4035-6648 at the 5' end (including the catalytic active center of histone methyltransferase - the SETdomain domain), the 5' and 3' primers are: 5'-cccgggCCAAGGTTGATTTCTGATAG-3'( SEQ ID No: 5), and 5'-gcggccgcCTAGCCTAGGAAGAC-3' (SEQ ID No: 6). The 50 μl PCR amplification system contained template pUC19-SDG701200ng, 20 pmol of 5' and 3' primers, 5 μl of 10×buffer, 20 μM of dNTP, and 1 μl of high-fidelity KODplus (TOYOBO). Amplification was performed under the following conditions: pre-denaturation at 94°C for 2 minutes; denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 68°C for 3 minutes, a total of 30 cycles. The PCR product was Sma I and not The restriction site I was cloned into the Escherichia coli expression vecto...
Embodiment 3
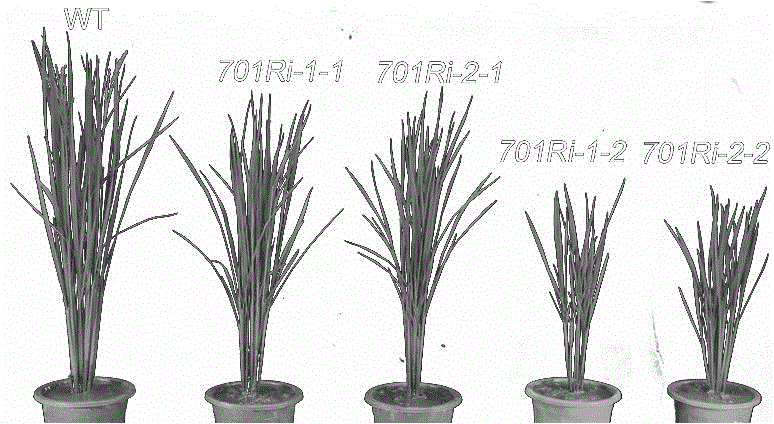
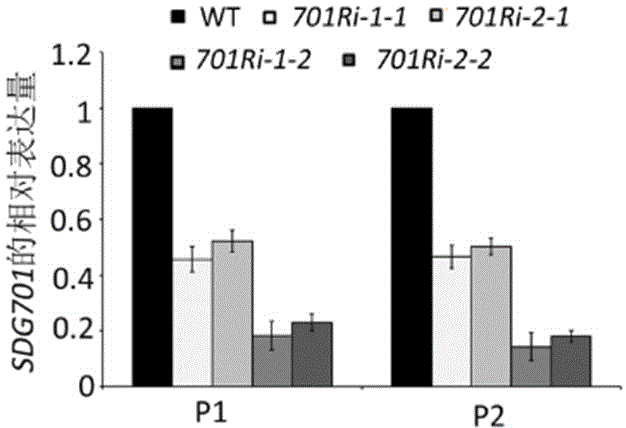
[0034] Example 3. Detection SDG701 Regulatory effect on rice growth and development
[0035] one, SDG701 Construction of antisense expression vector
[0036] Using RNA interference (RNAi) technology to SDG701 The gene is the target gene, and the RNA interference vector is constructed. Using the pUC19-SDG701 obtained in Example 1 as a template, PCR amplification was performed with two pairs of different primers SDG701 The nucleotide sequence from the 98th to the 463rd position of the 5' end is a complementary DNA duplex as a hairpin structure. The primer pair 1 used was: 5'-ccatggggatccGGGAGCTGCTGCTCAATGGGGAG-3' (SEQ ID No: 7) and 5'-gaattcCCGACGAGCCGAGCTTCTT-3' (SEQ ID No: 8), and the obtained PCR product was fSDG701i-1 (forward). Primer pair 2 is 5'-tctagaGGGAGCTGCTCAATGGGGAG-3' (SEQ ID No: 9) and 5'-aagcttCCGACGAGCCGAGCTTCTT-3' (SEQ ID No: 10), and the obtained PCR product is rSDG701i-1 (reversed). In order to facilitate subsequent vector construction, the 5' and 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com