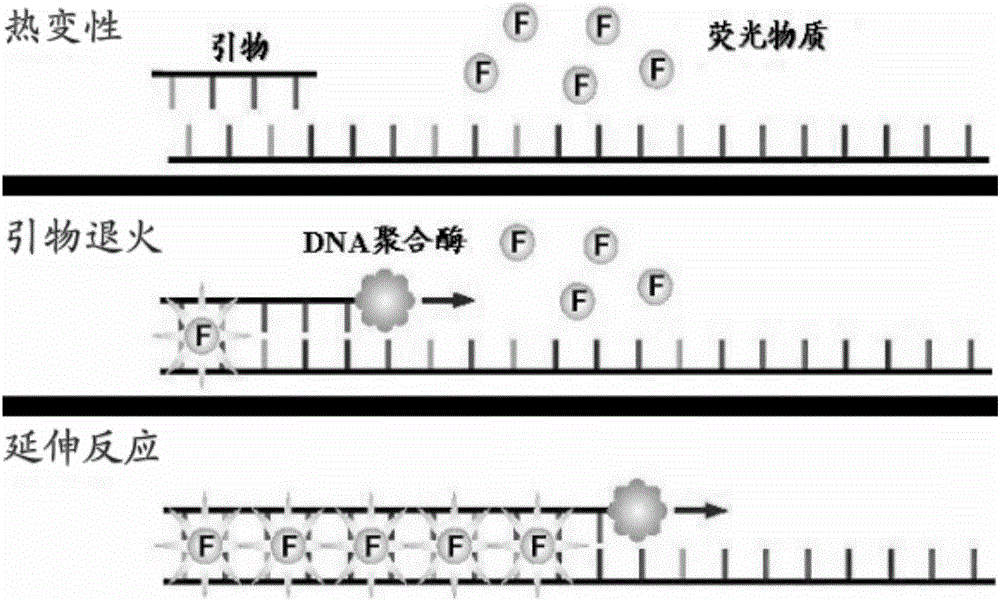
HPV (human papilloma virus) typing detection primers as well as detection method and application thereof
A technology of HPV66 and primer pairs, which is applied in biochemical equipment and methods, microbiological measurement/inspection, DNA/RNA fragments, etc., can solve the problems of inability to effectively apply clinical detection, low accuracy rate, and high quality requirements for personnel. Achieve rapid and accurate molecular detection methods, reduce inspection costs, and avoid cross-contamination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1: Primer design.
[0076] The primer-blast software on NCBI designed specific primers for different high-risk or low-risk HPV types to ensure that the primer pairs had no homology with other HPV types and human genomes. Afterwards, the obtained multiple pairs of primers (average 10 pairs) were input into the oligo 6 software for analysis, comprehensively considering the primer self-matching, neck loop structure, amplification efficiency and mismatching situation, and respectively obtained the optimal primers for different high-risk or low-risk types. Type HPV primer pair, as follows
[0077] The sequence of the high-risk HPV16-specific primer is shown in SEQ ID NO: 1-2:
[0078] SEQ ID NO: 1 (HPV16F): 5'---CAGACGACTATCCAGCGACC---3';
[0079] SEQ ID NO:2(HPV16R):5'---GCAGTGAGGATTGGAGCACT---3';
[0080] The sequence of the high-risk HPV18-specific primer is shown in SEQ ID NO: 3-4:
[0081] SEQ ID NO: 3 (HPV18F): 5'---CTGCTACACGACCTGGACAC---3';
[0082] SEQ ...
Embodiment 2
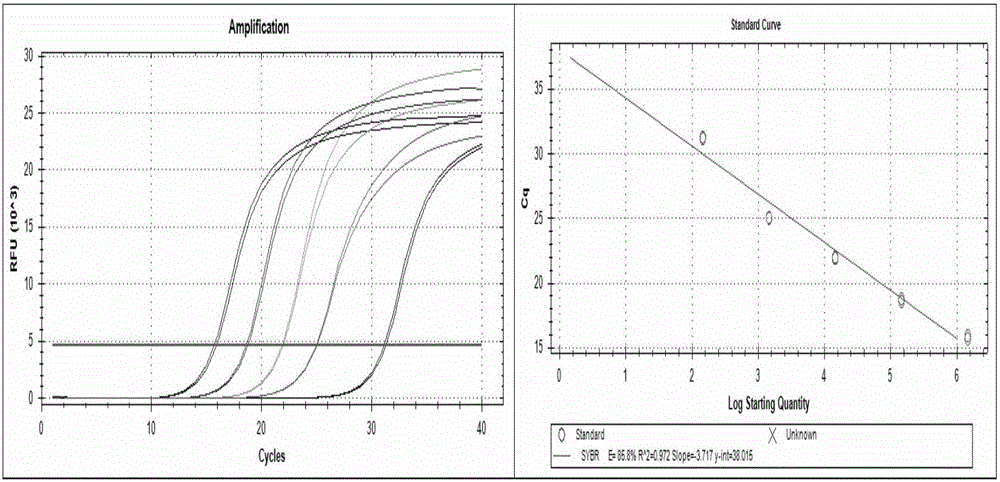
[0137] Example 2: Construction of plasmid standards and formulation of standard curves.
[0138] In order to test whether the designed primers can be accurately applied to the qPCR typing detection of HPV, the amplification standard curves of various primers are obtained correspondingly.
[0139] To obtain the standard curve of HPV53 as an example:
[0140] 1. Construction of 53-type plasmid standard:
[0141] (1) Target fragment amplification, using 53-type target fragment amplification primers (upstream: 5-GGTATCCGTATGGAGTGTG-3; downstream: 5-CCACCTTGCTACATACATGC-3), using HPV53-infected clinical sample DNA as a template. KOD-Plus-Neo (TOYOBO) kit was used for amplification, and the reaction system and reaction conditions are shown in Tables 1 and 2.
[0142] Table 1 Plasmid Construction Target Fragment Amplification System
[0143]
[0144] Table 2 Plasmid construction target fragment amplification reaction conditions
[0145]
[0146] (2) Gel recovery and purific...
Embodiment 3
[0162] Example 3: Detection of primer specificity.
[0163] In order to prove the amplification specificity of various specific primers to the target type, all specific primers in the invention were amplified with the DNA of clinical samples infected by 21 common individual HPVs, in which various clinical samples were tested by a third party to verify the infection Types of. And all have undergone positive reference amplification to ensure DNA concentration.
[0164] Now take type 16 primer as an example
[0165] 1. DNA extraction from clinical samples infected with 21 separate HPVs:
[0166] (1) Tighten the cap of the preservation tube containing the sample, and vortex for 15 seconds to suspend as much exfoliated cells on the cervical brush as possible in the preservation solution.
[0167] (2) Pipette 1ml of preservation solution into a 1.5ml EP tube, centrifuge at 8000g for 1 minute, and discard the supernatant.
[0168] (3) Use the protocol of QIAGEN (Germany) DNA MINI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com