Method and primer for fast detecting Cronobacter sakazakii at constant temperature and application
A technology of Cronobacter sakazakii, applied in the field of rapid constant temperature detection of Cronobacter sakazakii, which can solve the problems of lack of generality and specificity of primers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1-6
[0187] Embodiment 1-6 Cronobacter sakazakii constant temperature reaction system and detection method
[0188] Follow the steps (1) to (3) below for testing:
[0189] (1) Extraction of genomic DNA
[0190] The strains of Cronobacter sakazakii used for the detection were obtained from the China Center for the Collection of Industrial Microorganisms, numbered CICC21560 (=ATCC29544). Take 1mL of bacterial culture and use Beijing Tiangen Bioengineering Company's Bacterial Nucleic Acid Extraction Kit to extract genomic DNA, DNA OD 260 / OD 280 It was 1.8, and the concentration was 288ng / μL.
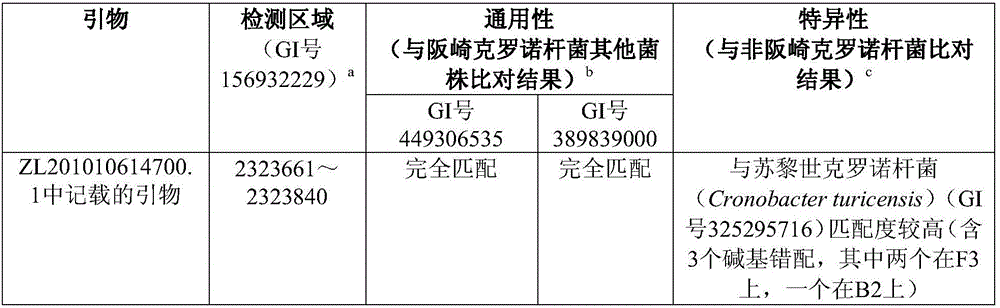
[0191] (2) Using the genomic DNA of Cronobacter sakazakii to be tested as a template, using self-prepared kits (see Table 2, Table 3) respectively, and preparing a reaction system according to the conditions described in Table 3, using Gram Sakazaki The Rhinobacter specific amplification primer set is used as primers for constant temperature amplification reaction. The primers in Examples ...
Embodiment 7
[0195] Example 7 Specific detection of Cronobacter sakazakii
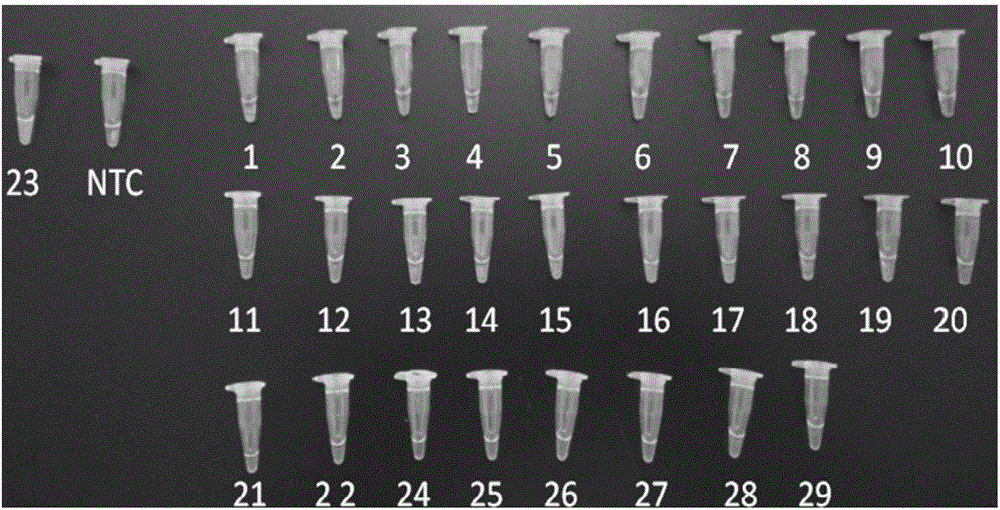
[0196] Collect 28 strains of non-Sakazakii Cronobacter (Table 4 and figure 1 1~22, 24~29 in ), these strains were compared with Cronobacter sakazakii strains (Table 4 and figure 1 23) in 23) are cultured respectively, get 1mL bacterium liquid, adopt kit 1A, extract bacterial DNA, and with reference to the reaction system and condition of embodiment 1, carry out LAMP amplification respectively (primer group is A) and add chromogenic agent to observe .
[0197] The test results are shown in Table 4 and figure 1 as shown, figure 1 Among them, 1-22 are Staphylococcus aureus, Staphylococcus aureus subsp. aureus, Staphylococcus epidermidis, Rhodococcus equi, Bacillus cereus, Bacillus mycoides, Listeria monocytogenes, Listeria innoculus Tertiary bacteria, Listeria evanii, Salmonella enterica subsp. enterica, Salmonella enteritidis, Salmonella typhimurium, Salmonella paratyphi B, Shigella dysenteriae, Shigella baumanni...
Embodiment 8
[0201] Embodiment 8 Sensitivity detection
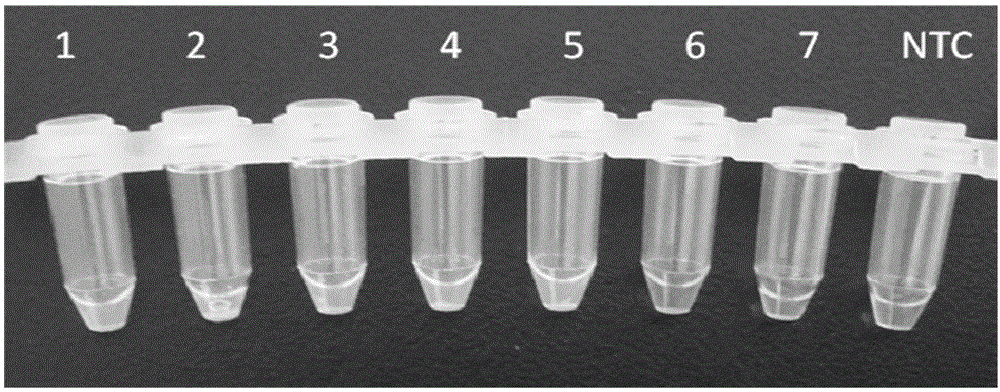
[0202] Extract the DNA of bacteria CICC 21560 according to the method of Example 2, adopt kit IIB, and add reaction system according to 50ng, 5ng, 500pg, 50pg, 5pg, 500fg, 50fg DNA gradient, other reaction conditions refer to the method of Table 3 Example 2 Carry out LAMP amplification (primer set is B') and add chromogen for observation respectively. Such as figure 2 As shown, 1-7 are 50ng, 5ng, 500pg, 50pg, 5pg, 500fg and 50fg respectively, NTC: negative control. figure 2 The reaction products treated with 50ng, 5ng, 500pg, 50pg, and 5pg were bright green, which was a positive result, and the reaction products treated with 500fg and 50fg and the negative control were orange, which was a negative result. The test results show that each reaction tube can still be detected when the minimum DNA content is 5pg (equivalent to about 1000 bacteria), and the sensitivity is high.
[0203] According to the above detection method, other s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com