Fluorescent quantitative PCR primer, probe, kit and detection method for detecting avian influenza subtype
An influenza virus, multiple fluorescence quantitative technology, applied in the field of bioengineering, can solve problems such as low sensitivity, achieve high sensitivity, shorten detection time and detection times, and avoid cumbersome operations.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0068] In the present invention, the preparation method of the positive plasmid standard substance preferably comprises the following steps:
[0069] A, extract the total RNA of H5, H7 and H9 subtype influenza virus samples respectively;
[0070] B. The total RNA of the H5, H7 and H9 subtype influenza viruses obtained in the step A is respectively reverse-transcribed to obtain the cDNA of the H5, H7 and H9 subtype influenza viruses;
[0071] C, using the cDNA obtained in step B as a template, using the nucleotide sequences shown in SEQ ID No.13 and SEQ ID No.14 as upstream primers and downstream primers to amplify H5 subtype influenza virus-specific target fragments , using the nucleotide sequences shown in SEQ ID No.15 and SEQ ID No.16 as upstream primers and downstream primers to amplify H7 subtype influenza virus-specific target fragments, using SEQ ID No.17 and SEQ ID No.18 The nucleotide sequences shown are respectively used as upstream primers and downstream primers to ...
Embodiment 1
[0109] Primer design
[0110] According to the M gene of AIV in GenBank and the hemagglutinin gene sequences of H5, H7 and H9 subtype avian influenza viruses, DNAMAN software was used for homology analysis, and Primer Express5.0 software was used to design specific primers and TaqMan in its conserved regions. fluorescent probe. Wherein, M gene primers and probes are obtained according to laboratory operation standards of avian influenza virus. The designed primer pairs and probes of H5, H7 and H9 subtype avian influenza viruses and M gene primer pairs and probes were entrusted to Shanghai Biological Engineering Company to synthesize.
[0111] RNA extraction
[0112] Take an appropriate amount of chicken embryo allantoic fluid, centrifuge at 12,000 rpm for 5 minutes, take 500 μl of supernatant, add 700 μl of Trizol to it, shake and mix, let stand for 10 minutes, and centrifuge at 12,000 rpm at 4°C for 15 minutes, take the supernatant and add it to 500 μl of isopropanol, mix w...
Embodiment 2
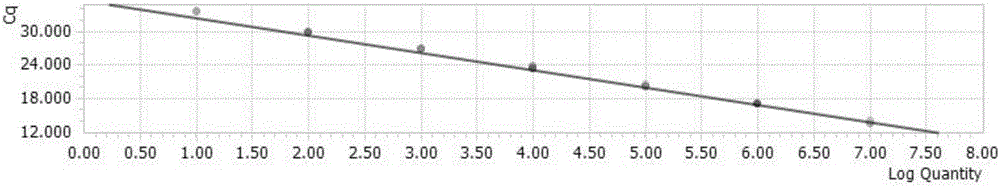
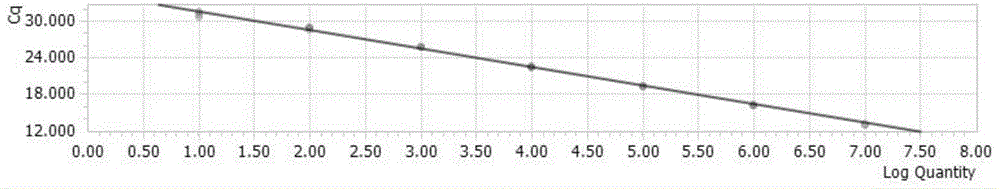
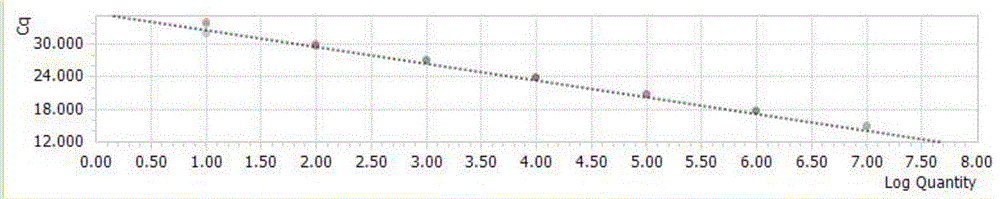
[0120] Dilute the recombinant plasmid of the universal vector according to a 10-fold gradient to obtain 1×10 0 Copy number / μl~1×10 7 Copy number / μl serially diluted plasmids were detected and analyzed in order to obtain the lower limit of detection. Obtain the Ct values of different concentrations of standard products to determine the minimum copy number of templates that can be detected by this method. At the same time, use the above-mentioned different concentrations of templates for detection by conventional PCR methods, and identify PCR products by 1% agarose gel electrophoresis. The amplification curve of each specific target fragment is as follows Figure 5-8 shown.
[0121] The study found that when the Ct value > 35 cycle is negative, and the Ct value ≤ 35 cycle is positive. The detection limit of the established fluorescent quantitative PCR method for detecting H5, H7, H9 influenza virus and influenza virus M gene was 10 copies.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com