GLP-1(7-37) polypeptide analog
1. GLP-1M, peptide technology, applied in the direction of peptides, hybrid peptides, specific peptides, etc., can solve the problems of increasing the risk of side effects of drugs, increasing the cost of taking drugs, and having a half-life of only 13 hours
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment l
[0044] Embodiment 1: the design of the GLP-1M gene of codon optimization, synthesis and expression plasmid construction
[0045] According to the designed protein sequence, use software to optimize the codon design of its polynucleotide sequence. After optimization, add the necessary sequence between promoter and rbs sequence, rbs sequence, rbs and start expression codon at the 5 end of its polynucleotide sequence ATG spacer sequence, ATG and six histidine codons and other sequences. The above sequence was entrusted to Shanghai Jierui Company to synthesize, and the synthetic gene was inserted into the expression vector pET24(+) between BamH1 / SalI to complete the construction of the expression plasmid. The sequence involved is as follows
[0046] Protein sequence GLP-1M(A8S / V33R)
[0047] H SEGTFTSDVS SYLEGQAAKE FIAWLRRGRG
[0048] Synthetic polynucleotide sequence for expressing SUMO-GLP-1M(A8S / V33R) fusion protein
[0049] ATTTTGTTTAACTTTAATAAGGAGATATACCATGCATCACCATCATCAC...
Embodiment 2
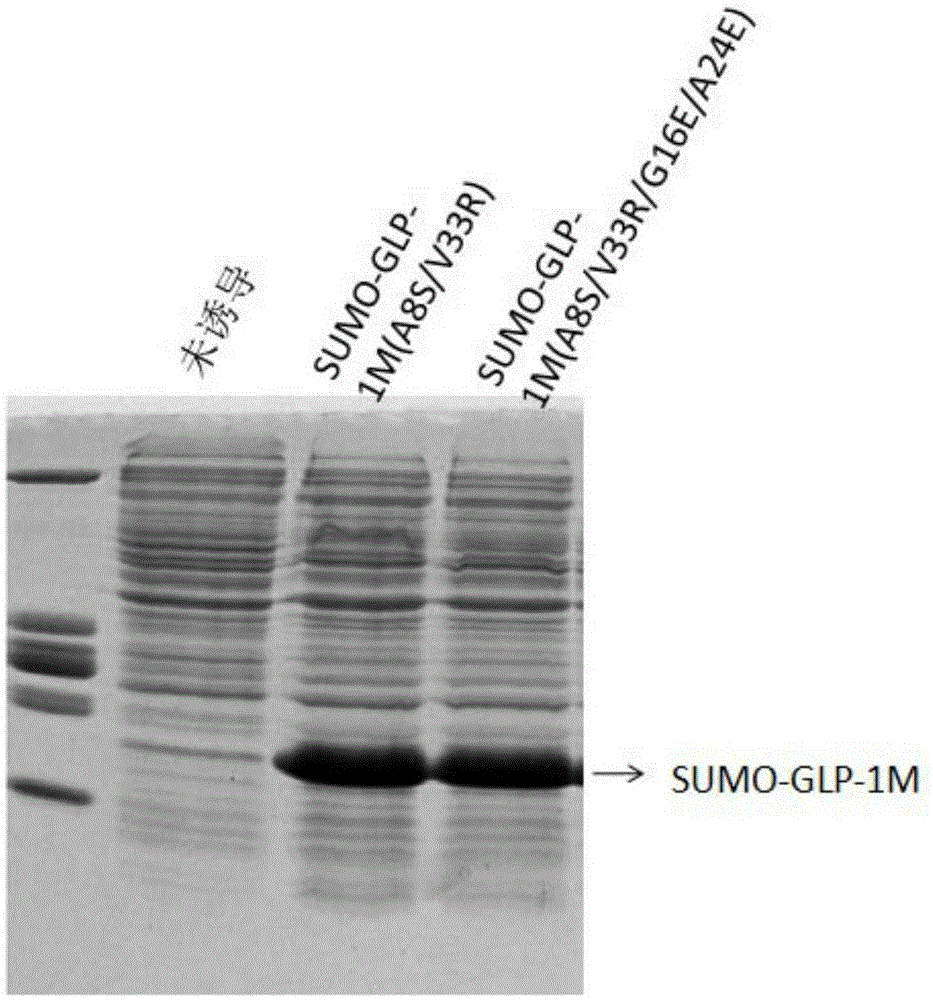
[0054] Example 2: SUMO-GLP-1M (A8S / V33R) and SUMO-GLP-1M (A8S / V33R / G16E / A24E) protein expression
[0055] The recombinant vector with correct sequencing results was transformed into Escherichia coli BL21 (DE3) (E. coli) host cells, and used as an engineered bacterium expressing recombinant proteins to express SUMO-GLP-1M protein. The engineering culture medium is 2×YT medium (10g / L tryptone; 5g / L yeast powder; 10g / L NaCl). Pick a single spot of bacteria containing the recombinant plasmid and place it in 10 ml of 2×YT medium, shake at 230 revolutions per minute (rpm), and culture overnight at 37°C. Transfer 5ml of overnight bacteria into 500ml of 2×YT liquid medium, shake and culture at 37°C until the recombinant engineered bacteria grow to OD600nm≈0.4~1, add IPTG at a final concentration of 0.2mM to induce, and carry out 16h recombinant protein at 30°C expression. see results figure 1
Embodiment 3
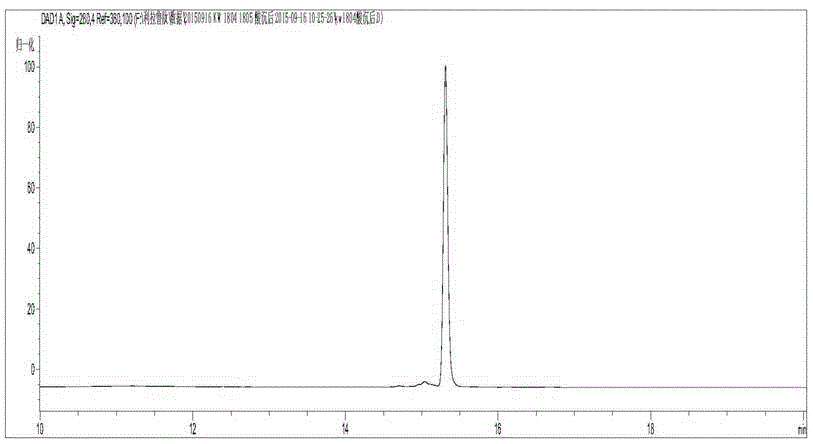
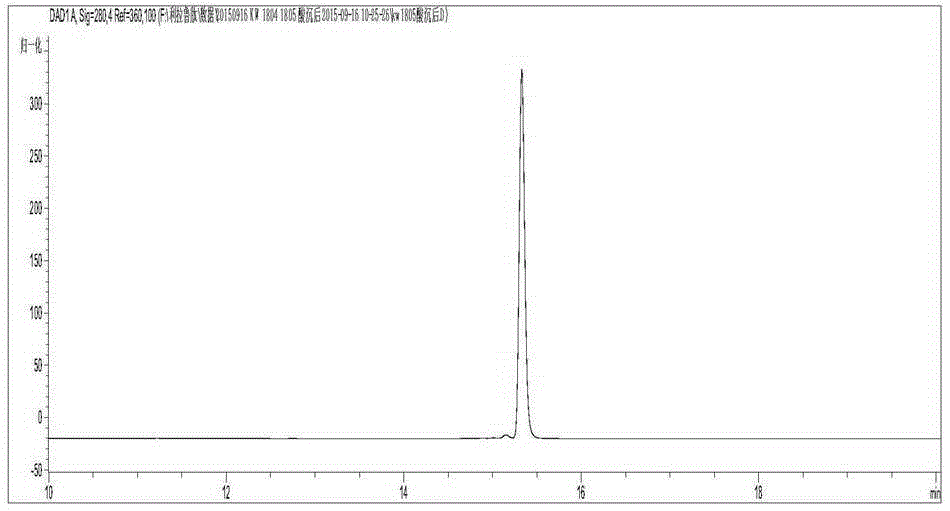
[0056] Example 3: Polypeptide purification, fatty acid modification and purification after modification
[0057] Cell collection and crushing: Centrifuge the fermentation culture, discard the supernatant, harvest the bacterial pellet, and weigh it; wash the pellet with buffer A (pH 8.0, 50mM PB, 500mM NaCl), and then resuspend it in buffer A for ultrasonication Crush, and then centrifuge the bacteriostasis solution with a high-speed centrifuge (16000 rpm, 30 min, 4° C.), and collect the supernatant.
[0058] Affinity chromatography and enzyme digestion: put 10ml of Ni affinity chromatography medium into the affinity column, equilibrate the chromatography column with buffer A, then load the sample, wash with Buffer L until no protein flows out, and the affinity is completed. The SUMO-GLP-1M fusion protein was collected by elution with Buffer A containing 300mM imidazole. Add SUMO protease at a mass ratio of 1:100 to digest the fusion protein overnight for 4 nights.
[0059] H...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com