Surface-enhanced Raman detection of biomolecules based on hyperbranched nanostructures
A surface-enhanced Raman and nanostructure technology, which is applied in Raman scattering, measuring devices, and analytical materials, can solve the problem that the amplification factor of molecular signals is difficult to achieve consistency, specific quantitative detection cannot be achieved, and molecular concentration cannot be obtained from molecular signals, etc. problem, to achieve the effect of low cost, elimination of competition, and good reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Preparation of hyperbranched nanoparticles:
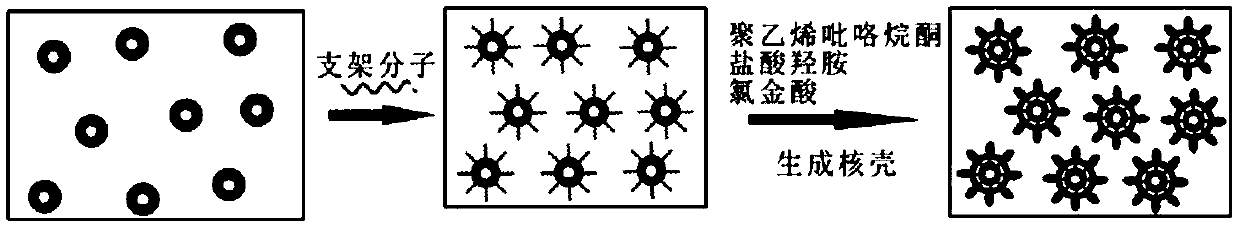
[0072] figure 1 A schematic diagram of the experimental procedure for the preparation of hyperbranched nanoparticles is given.
[0073] Taking gold core and gold shell as an example, the specific preparation method is:
[0075] 5'-AAAAAAAAAAAAAAAAAAAAACAAGAGTTACTAGTCTCGTCGGAGTCGTATCGCTACAAGTCC-3'
[0076] (1) First take 100ul 10nM 13nm gold balls, add 4ul 100uM scaffold DNA, mix and add 2ul 500mm
[0077] Sodium citrate solution is aged, after reacting for 15-30min, centrifuge and wash three times at 12000rpm for 15min to remove unassembled
[0078] The DNA is dispersed with a buffer solution of 100ul 10mM PB (0.1M NaCl, pH=7.4);
[0079] (2) The mixture is diluted with 900ul 0.1M PBS (pH=7.4) buffer solution, and set aside;
[0080] (3) Take 100ul of the solution in (2), then add 50ul 1% PVP, 20ul 10mM NH 2 OH-HCl, repeat the above operation six times, and finally add 4ul, 8ul, 12ul, 16ul, 20ul...
Embodiment 2
[0082] Specific ultrasensitive quantitative detection of DNA
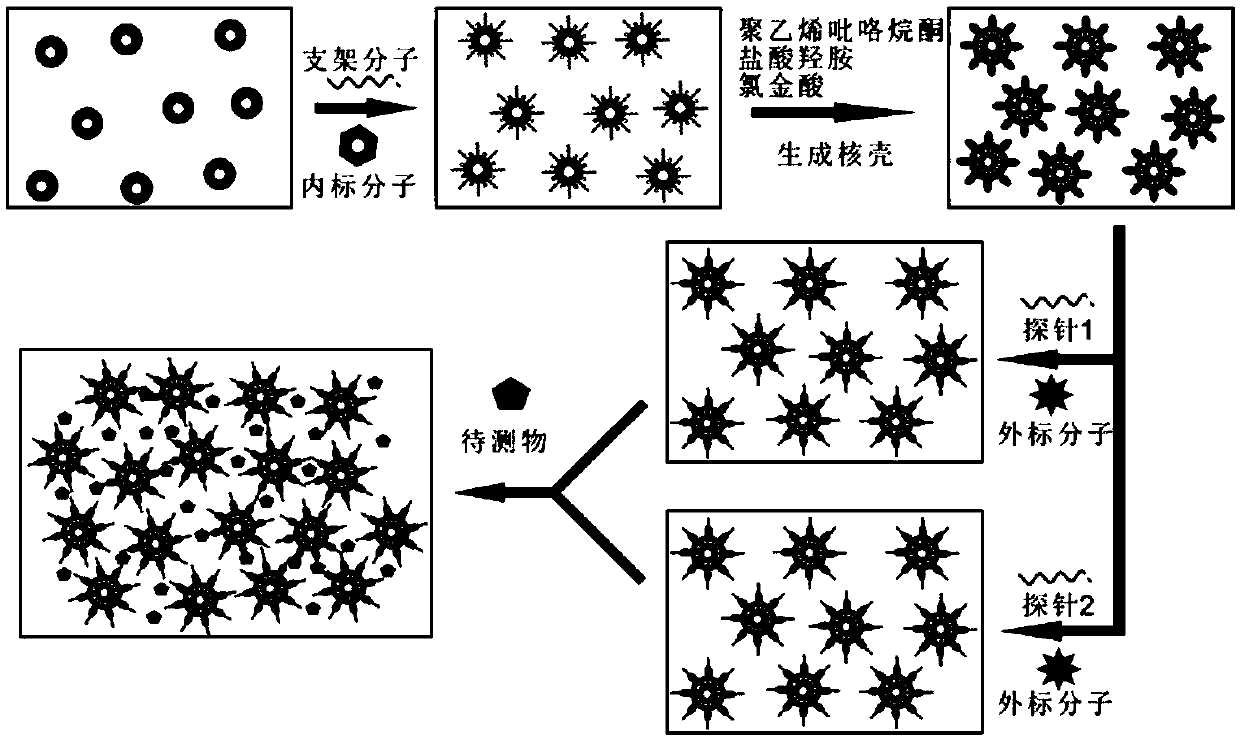
[0083] image 3 The flow chart of specific quantitative detection of DNA is given.
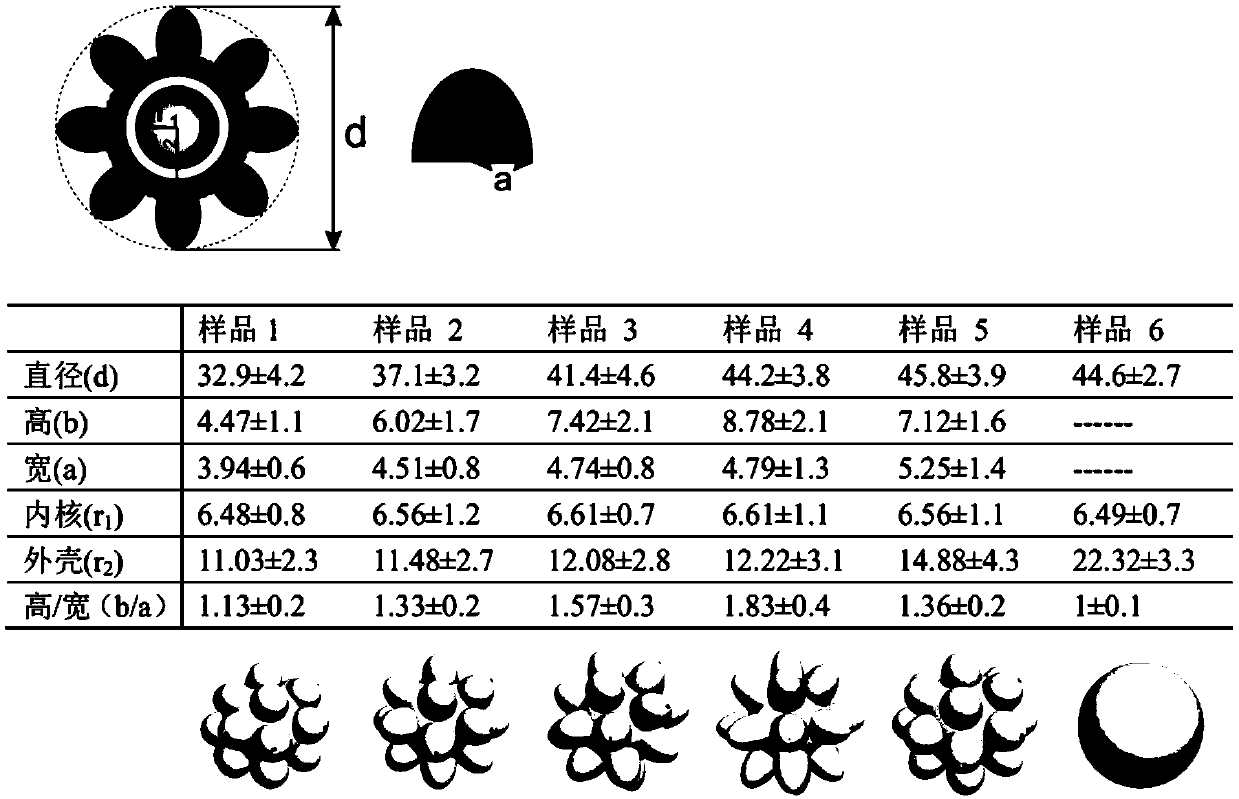
[0084] by figure 2 Middle No. 4 (sample 4) hyperbranched nanoparticles were used as SERS probes, 4-MBA was used as internal standard, and DTNB was used as external
[0085] Mark as an example:
[0086] Scaffold DNA:
[0087] 5'-AAAAAAAAAAAAAAAAAAAAACAAGAGTTACTAGTCTCGTCGGAGTCGTATCGCTACAAGTCC-3'
[0088] Probe 1: 5'-AAAAAAAAAA TTTTT ATgATgTTCg TTgTg-3'
[0089] Probe 2: 5'-gTgTT TAggATTTgC TTTTT AAAAAAAAAA-3'
[0090] DNA to be tested: 5'-gCAAA TCCTAAACAC CACAA CgAAC ATCAT-3'
[0091] 1. Preparation of hyperbranched nanostructured SERS probes
[0092] (1) First take 100ul 10nM 13nm gold balls, add 4ul 100uM scaffold DNA, mix and add 2ul 500mm sodium citrate solution for aging, after reacting for 15-30min, centrifuge and wash three times at 12000rpm for 15min, remove unassembled DNA, and use 100ul 10mM PB (0.1M NaCl, pH=7.4)...
Embodiment 3
[0103] Specifically recognizes RNAs of the let 7 family
[0104] image 3 A flowchart for the specific recognition of RNAs of the let 7 family is given.
[0105] by figure 2 The No. 4 hyperbranched nanoparticles are used as SERS as probe, 4-MBA as internal standard, and DTNB as external standard as an example:
[0106] Scaffold DNA:
[0107] 5'-AAAAAAAAAAAAAAAAAAAAACAAGAGTTACTAGTCTCGTCGGAGTCGTATCGCTACAAGTCC-3'
[0108] Probe 1: 5'-AAAAAAAAAAAAACTATGCAA-3'
[0109] Probe 2: 5'-CCTACTACCTCTAAAAAAAAAA-3'
[0110] let 7a:5'-UGAGGUAGUAGGUUGUAUAGUU-3'
[0111] let 7b:5'-UGAGGUAGUAGGUUGUGUGGUU-3'
[0112] let 7c:5'-UGAGGUAGUAGGUUGUAUGGUU-3'
[0113] let 7d:5'-AGAGGUAGUAGGUUGCAUAGUU-3'
[0114] let 7e:5'-UGAGGUAGGAGGUUGUAUAGUU-3'
[0115] let 7f:5'-UGAGGUAGUAGAUUGUAUAGUU-3'
[0116] let 7g:5'-UGAGGUAGUAGUUUGUACAGUU-3'
[0117] let 7i:5'-UGAGGUAGUAGUUUGUGCUGUU-3'
[0118] miR 98:5'-UGAGGUAGUAAGUUGUAUUGUU-3'
[0119] mirR 21:5'-UAGCUUAUCAGACUGAUGUUGA-3'
[0120] 1. Prepa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com