Specific nucleic acid aptamer and application
A nucleic acid aptamer and species-specific technology, which can be used in DNA/RNA fragments, measuring devices, instruments, etc., and can solve the problems of long time, cumbersome operation steps, and complicated operations.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
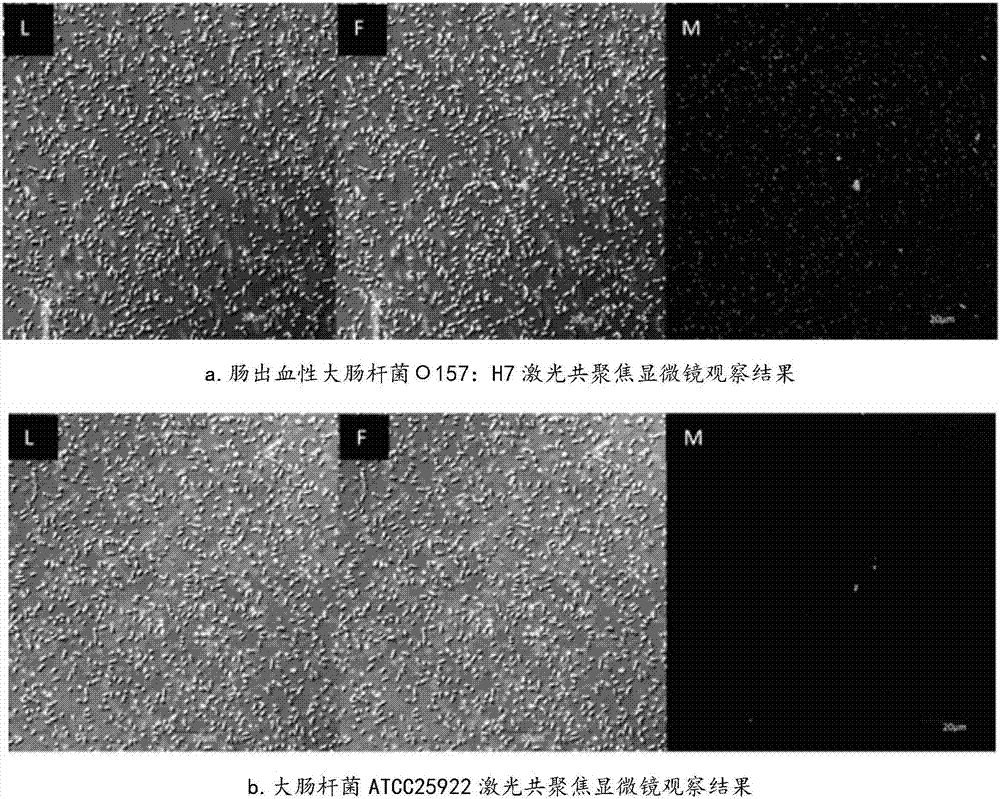
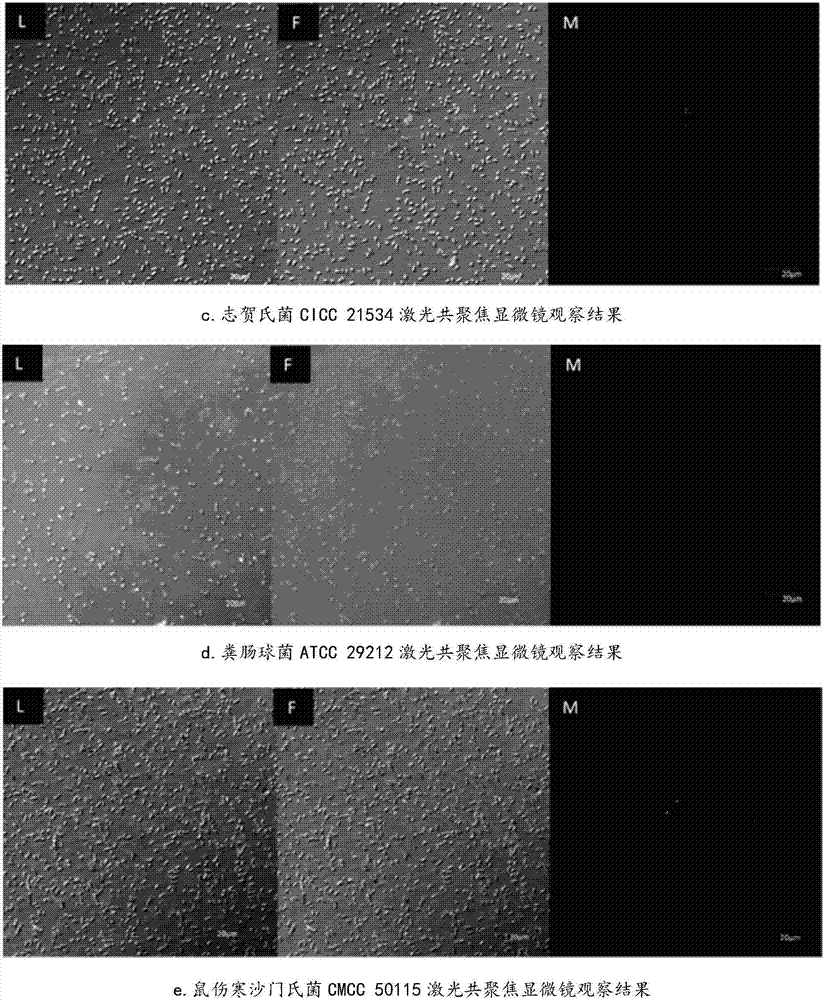
[0024] Screening of nucleic acid aptamers specifically binding to enterohemorrhagic Escherichia coli O157:H7 by whole-bacteria subtractive SELEX technology
[0025] (1) Synthesis of random single-stranded DNA (ssDNA) library and primers
[0026] Random ssDNA library references are as follows:
[0027] 5'-GCAATGGTACGGTACTTCC-N45-CAAAAGTGCACGCTACTTTGCTAA-3', the middle random region is 45 nucleotides, both ends are fixed regions, the total sequence length is 88 nucleotides, and the library capacity is 4 45 . The primer sequences used for screening are:
[0028] Primer Ⅰ: 5'-GCAATGGTACGGTACTTCC-3'
[0029] Primer Ⅱ: 5'-TTAGCAAAGTAGCGTGCACTTTTG-3'
[0030] Primer Ⅲ: 5'-GCTAAGCGGGTGGGACTTCCTAGTCCCACCCGCTTAGCAAAGTAGCGTGCACTTTTG-3'
[0031] Among them, Primer I needs FITC fluorescent group to be modified during the preparation of the secondary library. In the constructed fluorescence detection method, aptamer A46 needs to be modified with biotin.
[0032] The above nucleic aci...
Embodiment 2
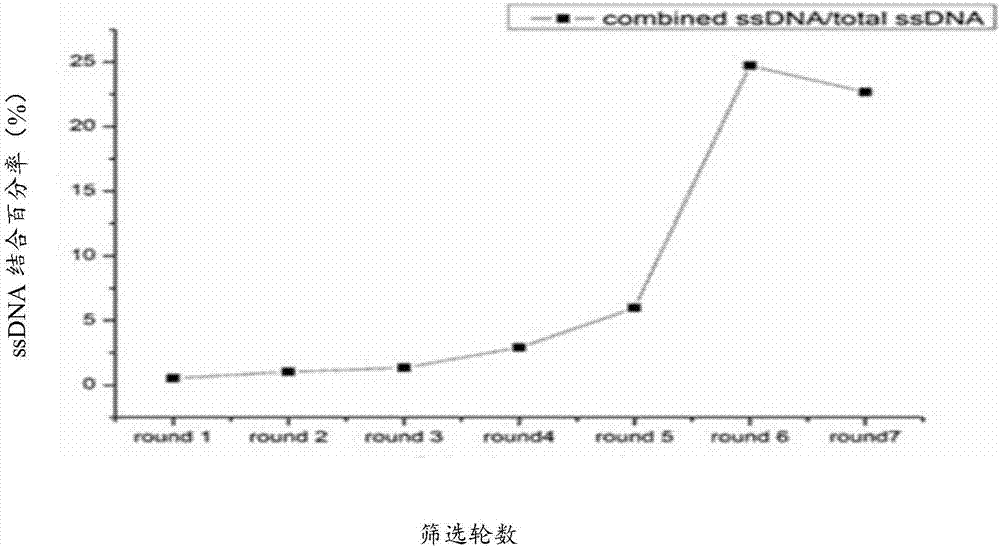
[0057] Fluorochemical analysis to monitor the screening process
[0058] (1) Preparation of fluorescent primary library and secondary library
[0059] 1) The initial random ssDNA library labeled with FITC fluorescence was directly synthesized by Shanghai Sangon Biotechnology Co., Ltd.;
[0060] 2) The FITC fluorescently labeled secondary ssDNA library was obtained by gel-cutting recovery and purification experiments, and the specific steps were as follows:
[0061] ①Using the ssDAN-target bacteria obtained in each round of screening as a template, the FITC-labeled Primer Ⅰ synthesized by Sangon as the upstream primer, and other primers and PCR conditions unchanged, refer to the PCR amplification system and PCR amplification system of the first round of screening in Example 1. condition;
[0062] ② Store the amplified product in the dark, and prepare the FITC-labeled ssDNA library according to the steps for preparing the secondary library in Example 1;
[0063] ③The FITC flu...
Embodiment 3
[0071] Cloning and Sequencing
[0072] (1) After 7 rounds of screening, the sixth round of screening product with the highest binding efficiency to the target bacteria was selected as a template, and the upstream primer Primer I and the downstream primer Primer II were used for PCR amplification. The amplification system and conditions were referred to in Example 1. PCR program;
[0073] (2) Connect the PCR product to the cloning vector pMD19-T, select blue and white spots, extract the plasmid after cell culture, and send the plasmid to Shanghai Sangon Biotechnology Co., Ltd. for sequencing;
[0074] (3) Use Chromas and DNAMAN software to perform homology and secondary structure prediction on the sequencing results, and finally obtain the sequence of the nucleic acid aptamer (named A46) specifically binding to Escherichia coli O157:H7 (sequence list 1) and predicted secondary structure diagram (see figure 2 ). Sequence length of aptamer A46 specifically binding to Escheri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com