Probe set, kit and method for rapid detection of BCR/ABL gene fusion
A gene fusion and probe set technology, applied in the field of molecular biology, can solve the problems of unfavorable signal confirmation and statistics, reduction of fluorescence signal intensity, divergence of fluorescence signal points, etc., to reduce non-specific hybridization, avoid non-specific hybridization, speed quick effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Design and synthesis of three kinds of probes: contact probe, amplification probe and fluorescent probe
[0042] (1) Through sequence comparison and advanced structural analysis, the sequences of 30 bp on the BCR gene and the ABL gene were respectively selected as the detection target site 1 and the detection target site 2, and their base sequences were respectively:
[0043] Target site 1: 5'-GCTACCGTTTCCAGTAGTTATTCCCCCTCCA-3'
[0044] Target site 2: 5'-TCCCCCTCCATCAGGCAGTTTCCCAGACAT-3'
[0045] (2) According to the sequence of the target site, design a complementary probe sequence, and add 20 groups of 20-base repeat sequences at its 3' end to form contact probe 1 and contact probe 2, and their base sequences are respectively:
[0046] Contact probe 1:
[0047] 5'-TGGAGGGGGATAACTACTGGAAACGGTAGC (GTATJCGCJCTGFTATJCCG) 20 -3'
[0048] Contact Probe 2:
[0049] 5'-ATGTCTGGGAAACTGCCTGATGGAGGGGGA(AGTFAJCGCFGTAFCAAJTJ) 20 -3'
[0050] The repetitive sequen...
Embodiment 2
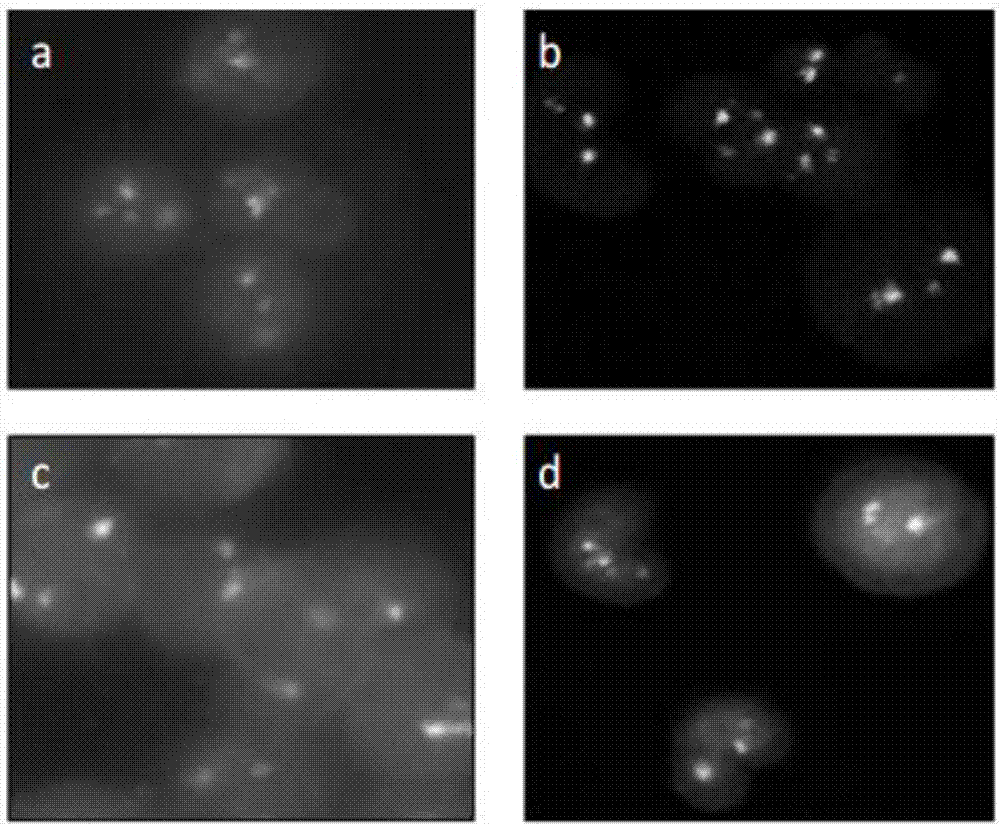
[0063] Embodiment 2: traditional FISH probe and probe set of the present invention detect BCR / ABL gene fusion
[0064] A. Traditional FISH probe detection
[0065] A method for detecting BCR / ABL gene fusion using traditional FISH probes, comprising the following steps:
[0066] (1) Take 10 μL of sample and drop it on the glass slide, and dry it at 56°C to fix the sample on the glass slide;
[0067] (2) Dewax the slides by immersing them in xylene for 2 times at room temperature, each time for 10 minutes, then immerse them in 100% ethanol for 5 minutes, then immerse them in 100% ethanol, 85% ethanol and 70% ethanol for 2 minutes each. water, immerse in purified water at 90°C for 20-30min, take out the glass slide for observation every 5min, to prevent the sample from falling off, take out the glass slide, and immerse it in 70% ethanol, 85% ethanol and 100% ethanol in sequence at room temperature Dehydrate in ethanol for 2 minutes each, and dry naturally;
[0068] (3) Add 3 μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com