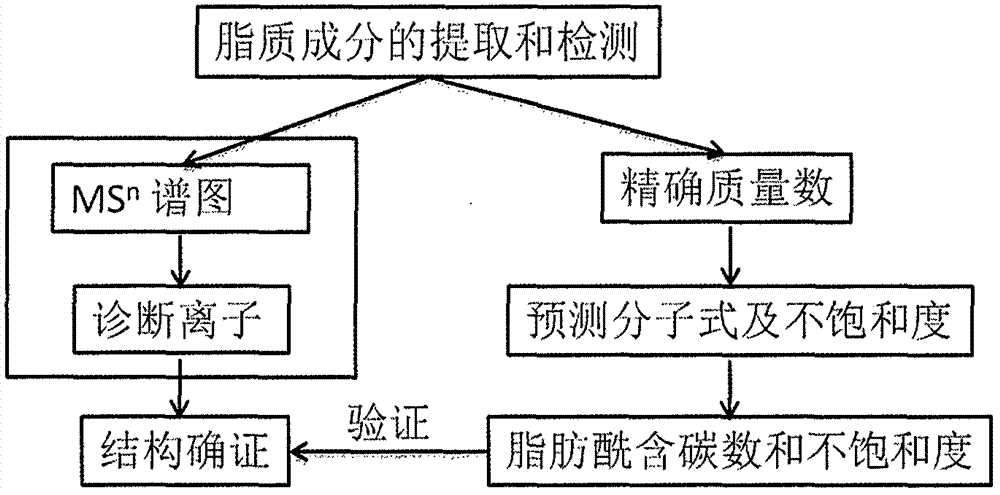
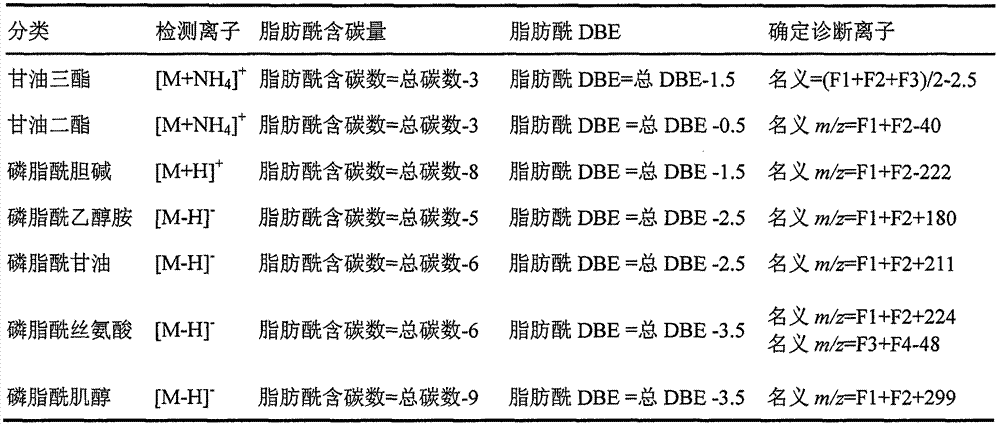
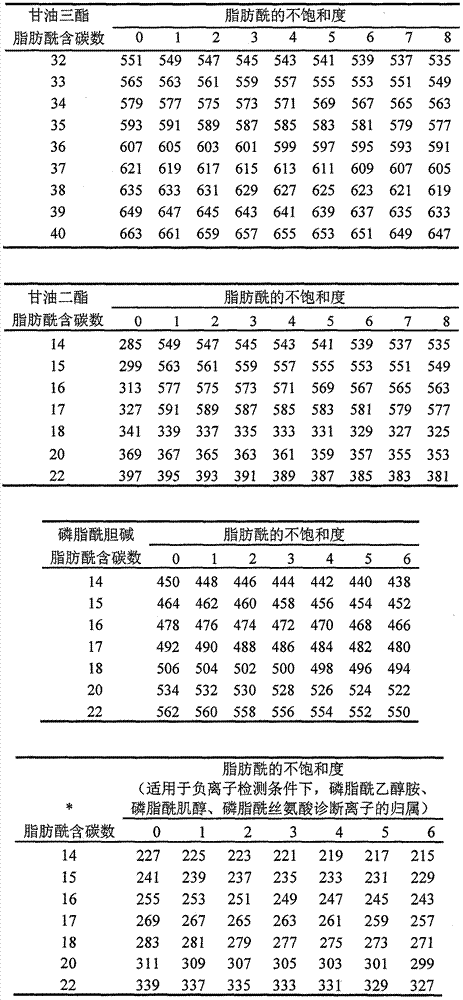
Method for identifying lipid component structure in lipidomics based on CID fragmentation
A technology of lipid composition and lipidome, applied in the field of life sciences, which can solve problems such as difficulties and bottlenecks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Composition Determination of Triglycerides in Human Plasma Samples
[0022] a: Extraction of lipid components in plasma samples
[0023] Select the Folch extraction method to extract triglycerides in the plasma sample, as follows: add 180 μL methanol to 20 μL plasma sample and vortex fully, then add 400 μL chloroform, vortex the sample, and finally introduce 150 μL water layer. After sufficient extraction, centrifuge at 8000 g for 10 min at 4°C. Remove the lipid-rich chloroform layer. The chloroform solvent was evaporated to dryness. Reconstitute with isopropanol / methanol solvent. Centrifuge and take the supernatant for injection.
[0024] b: Select the appropriate detection method and detection conditions
[0025] Chromatographic separation conditions: Chromatographic column: Hypersil GOLD aQ column (2.1×150mm, 3μm I.D.). Mobile phase: A is acetonitrile / water solution containing 0.1% formic acid and 5mM ammonium formate (40:60, V / V), B is isopropanol / acetonitrile...
Embodiment 2
[0030] Determination of phosphatidylcholine in lung tissue samples of SD rats
[0031] a: Extraction of lipid components in liver tissue samples
[0032] Select the methyl tert-butyl ether-methanol extraction method (MTBE extraction method) to extract the components of phosphatidylcholine in the liver tissue sample, as follows: After the 50 mg liver sample is homogenized, add 400 μL of pre-cooled 75% methanol solution, vortex After vortexing for 10 minutes, add 1 mL of methyl tert-butyl ether, vortex for 20 minutes, then add 60 μL Milli-Q ultrapure water, vortex for 10 minutes, let stand for 10 minutes, centrifuge at 17,000 rpm for 10 minutes, separate the samples, and take 900 μL of the supernatant and centrifuge The concentrator was evaporated to dryness. The upper layer concentrate was redissolved in 100 μL (isopropanol:methanol=8:2) mixture, sonicated in a water bath for 10 minutes, centrifuged at 17,000 rpm for 10 minutes, and 40 μL of the supernatant was taken for testi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com