Method for guiding helicobacter pylori to eradicate drug use based on PGM high throughput sequencing technology
A Helicobacter pylori, technical guidance technology, applied in the field of Helicobacter pylori drug resistance gene and host drug metabolizing enzyme CYP2C19 gene, can solve the problems of long detection cycle, easy pollution, low specificity, etc., achieve fast detection results, avoid The effect of waste and simple operation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
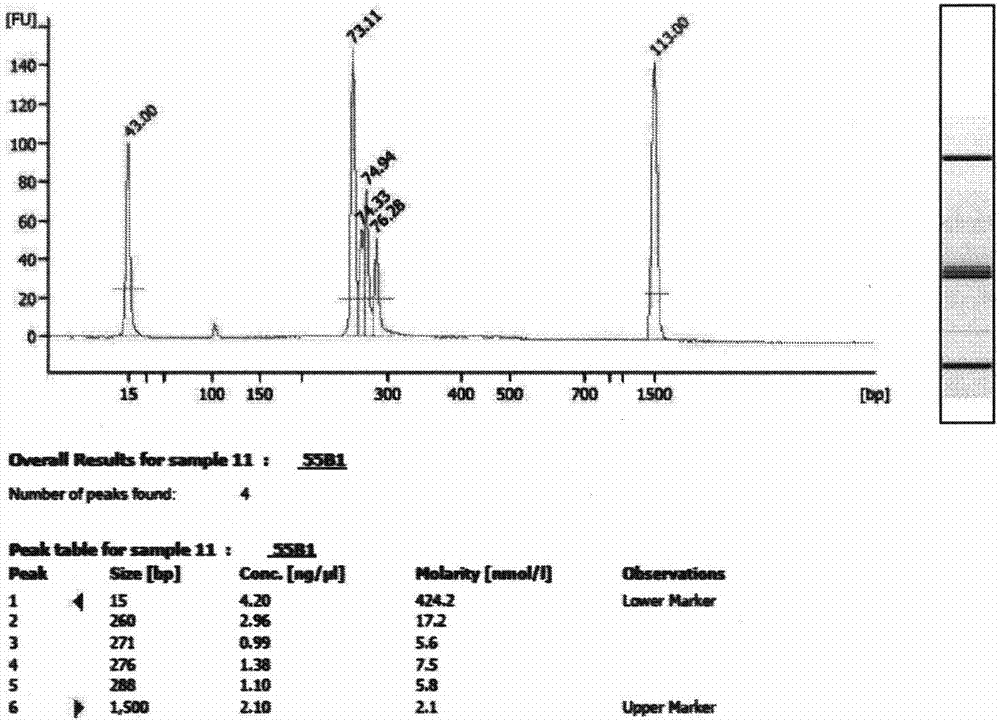
Image
Examples
Embodiment 1
[0022] Design specific primers covering VacA, 23SrRNA, gyrA, CYP2C19*2, and CYP2C19*3 gene regions to be tested. The amplified product is 180-220bp. See Table 1 for details.
[0023] Table 1 Specific primers for gene regions to be tested
[0024]
[0025]
[0026] A GCGTGTCTCCGACTCAG-bracode-GAT-specific primer was added to the 5' end of the forward primer of the primers designed above, and a CCTCTCTATGGGCAGTCGGTGAT-specific primer was added to the 5' end of each reverse primer. The sequence of barcode1 is CTAAGGTAAC. Different samples must use different barcode sequences to distinguish between samples. Other barcode sequences can be queried on the official website of Life Technologies. See Table 2 for details, where the red bold part is the barcode1 sequence.
[0027] Table 2 primers containing barcode1 sequence
[0028]
[0029] 3. The mixing ratio of these primers in the Primer Pool is: 23SrRNA:CYP2C19*2:CYP2C19*3:gyrA:VacA=2:1:1:1.2:1.5.
Embodiment 2
[0031] Genome extraction from gastric mucosa
[0032] (1) In a sterile 2ml round bottom EP tube, add 100μl tissue lysate and a sterile steel ball with a diameter of 3mm;
[0033] (2) Take out the gastric mucosal tissue from the sample preservation solution, and transfer it to a 2ml round-bottomed EP tube containing the lysate and steel beads;
[0034] (3) The tissue grinder grinds the sample, and the parameters are 50Hz and 60s at room temperature;
[0035] (4) After fully grinding, briefly centrifuge, and transfer the homogenate liquid to a new sterile 1.5ml EP tube;
[0036] (5) Incubate the 1.5ml EP tube containing the homogenized tissue in a 95°C metal bath for 10 minutes, and invert it several times in the middle;
[0037] (6) Quickly incubate EP in a refrigerator at 4°C for 30 minutes;
[0038] (7) Take out the EP tube from the refrigerator, centrifuge at 12,000 rpm for 5 minutes, collect the supernatant into a new EP tube, and then obtain the whole genome of the host a...
Embodiment 3
[0040] Amplification and purification of target fragments
[0041] (1) Prepare the following reaction system with PCR primers mixed according to the ratio: total reaction system 20 μl, including 10 μl of 2*Phusion HF Buffer PCR Master mix, 2 μl of template, 2 μl of primers, ddH 2 O 6 μl. The reaction conditions were pre-denaturation at 95°C for 3 minutes, denaturation at 95°C for 30 seconds within the cycle, annealing at 60°C for 30 seconds, extension at 72°C for 30 seconds, 18 cycles, and extension at 72°C for 5 minutes at the end of the cycle.
[0042] (2) Add 24 μl Agencourt AMPure XP beads magnetic beads to 20 μL of the amplification product, mix by pipetting, and place at room temperature for 5 minutes. When the liquid becomes clear, discard the supernatant, slowly add 200 μl of 80% ethanol to wash, discard the ethanol after 30 seconds, and repeat once. After the ethanol volatilized, 40 μl of water was added for elution when the center of the magnetic beads began to cra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com