Cymbidium sinense chlorophyll catabolism regulatory protein as well as coding gene and application thereof
A technology for chlorophyll degradation and metabolism regulation, which is applied in orchid chlorophyll degradation metabolism regulation protein and its coding gene and application field, which can solve the problems of long cycle and unpredictable traits, and achieve the effect of chlorophyll reduction and trait improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
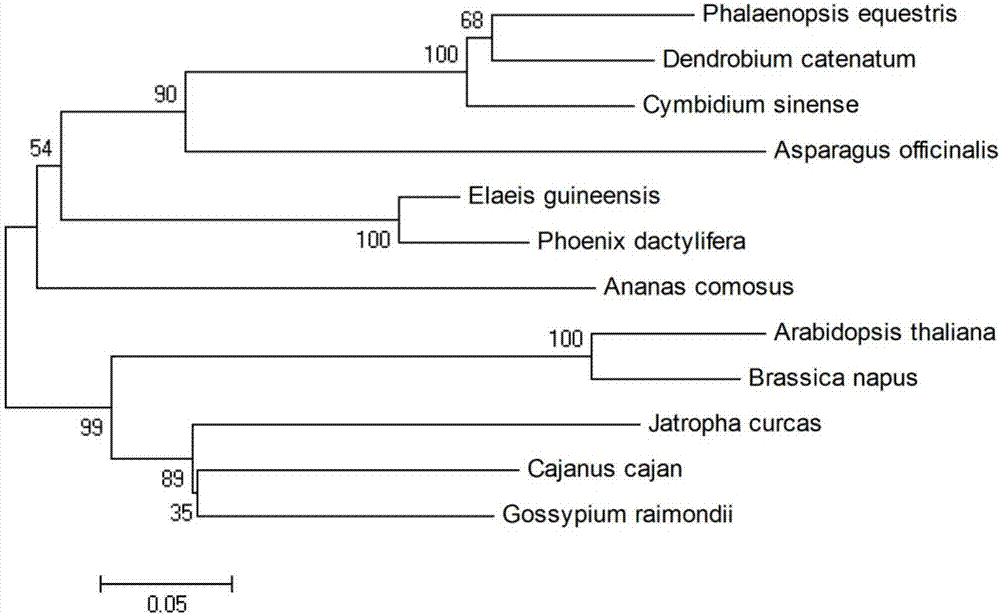
[0019] Example 1 Cloning and sequence analysis of CsCLH2 gene
[0020] 1. Extraction of RNA
[0021] Take 2 g of the young leaf tissue of Molan cultivar Dharma, extract its total RNA with plant Trizol (Invitrogen) reagent, and reverse transcribe it into cDNA (Thermo Scientific RevertAid First Strand cDNASynthesis Kit).
[0022] 2. Acquisition of the target gene CsCLH2
[0023] Using CsCLH2-F1 (SEQ ID NO: 3) and CsCLH2-R1 (SEQ ID NO: 4) as primers and the cDNA obtained in the above step as a template, PCR was performed using Ex-Taq enzyme (TaKaRa Biotechnology Co.) according to the following conditions: 94°C for 4min, then 34cycles (94°C for 40s, 59.5°C for 40s, 72°C for 1.5min, 72°C for 10min). The PCR product was recovered from the agarose gel, then connected to the pMD19-T vector (TaKaRa Biotechnology Co.) and sent to Huada Gene Research Institute for sequencing. The analysis of the sequencing results found that the amplified fragment contained the complete CDs sequence o...
Embodiment 2
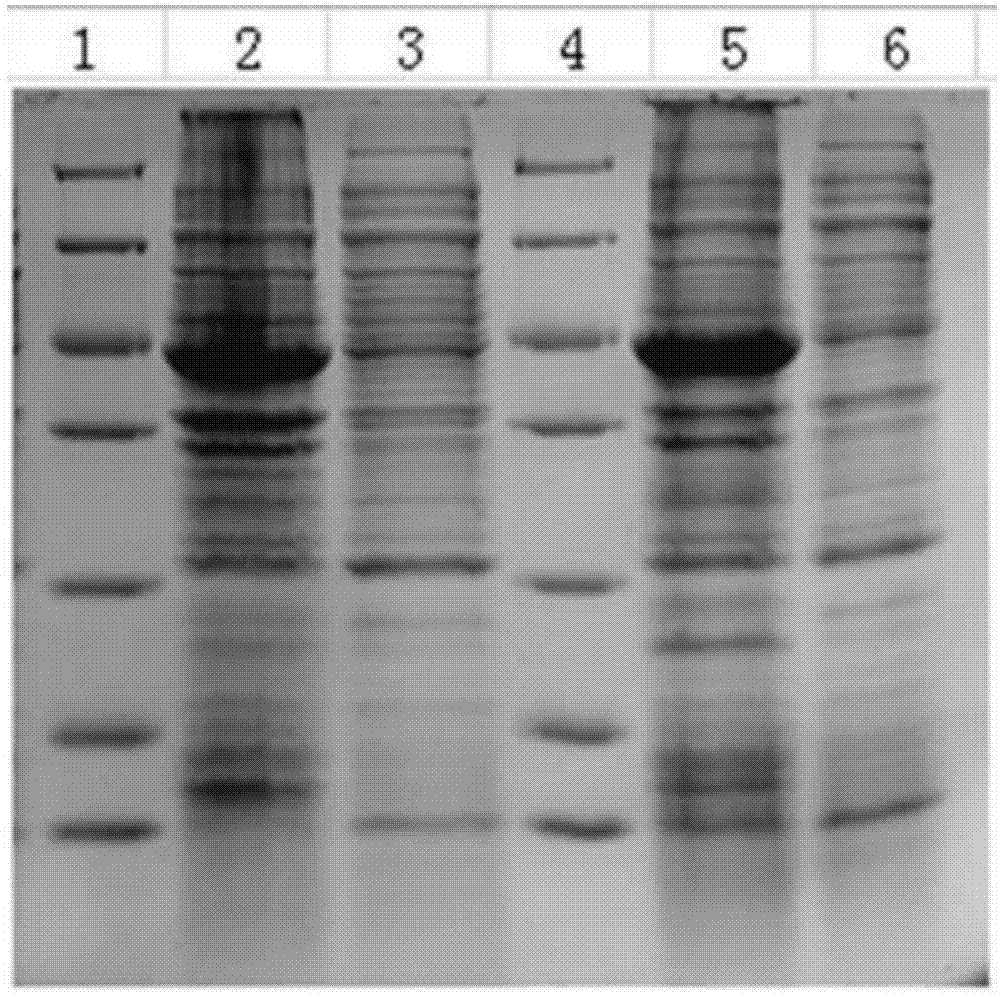
[0026] Example 2 Prokaryotic expression of CsCLH2 protein
[0027] 1. Protein prokaryotic expression vector construction
[0028] Using CsCLH2-F2 (SEQ ID NO: 5) and CsCLH2-R2 (SEQ ID NO: 6) as primers, using the cDNA of Molan leaves as a template, use Ex-Taq enzyme (TaKaRa Biotechnology Co.) to perform PCR according to the following conditions : 94°C for 4min, then 34cycles (94°C for 40s, 59.5°C for 40s, 72°C for 1.5min, 72°C for 10min). The PCR product (CsCLH2 gene) was recovered from the agarose gel, then connected to the pGEX6P-1 vector (TaKaRa Biotechnology Co.) and sequenced to verify its accuracy, confirming that the CsCLH2 gene was inserted into the pGEX6P-1 vector, thus obtaining pGEX6P- 1-CsCLH2 expression vector. The Escherichia coli containing pGEX6P-1-CsCLH2 is currently preserved in the Institute of Environmental Horticulture, Guangdong Academy of Agricultural Sciences.
[0029] 2. Protein induction expression and enzyme activity analysis
[0030] 1) Inoculate...
Embodiment 3
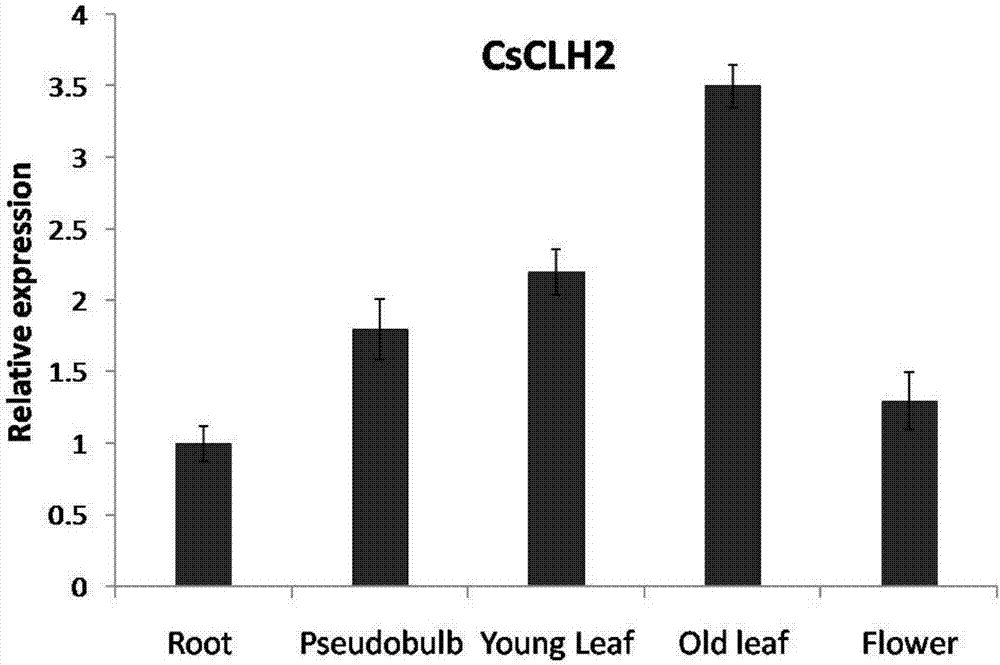
[0036] Example 3 Expression pattern of CsCLH2 in orchids
[0037] 1. Extraction of RNA
[0038] Take 2 g of different tissues of the young leaves, old leaves, roots, pseudobulbs, and flowers of Molan cultivar Dharma, use the plant Trizol (Invitrogen) reagent to extract its total RNA, and take 2 μl of it using the reverse transcription kit Thermo Scientific RevertAid First Strand cDNASynthesis Kit reverse transcription into cDNA.
[0039] 2. Quantitative PCR
[0040] Using primers CsCLH2QRT-F (SEQ ID NO: 7) and CsCLH2QRT-R (SEQ ID NO: 8), the expression of CsCLH2 gene in different tissues of orchids was detected by real-time quantitative PCR. Actin QRT-F (SEQ ID NO:9) and ActinQRT-R (SEQ ID NO:10) were used as primers to amplify Actin as an internal reference. The following procedure was followed: pre-denaturation at 95°C for 30 s, followed by 40 cycles (10 s at 95°C, 10 s at 59.5°C, 30 s at 72°C), and extension at 72°C for 10 min. The iCycler IQ Real-timePCR Detection Syst...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com