Method for determining DNA extraction rate of soil sample
A soil sample and extraction rate technology, applied in the field of soil biochemistry, can solve problems such as low comparability, bias in microbial information, and inability to measure DNA concentration, and achieve the effects of simple preparation, simple culture method, and quick and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1, a kind of method for measuring all soil sample DNA extraction rates, carries out following steps successively:
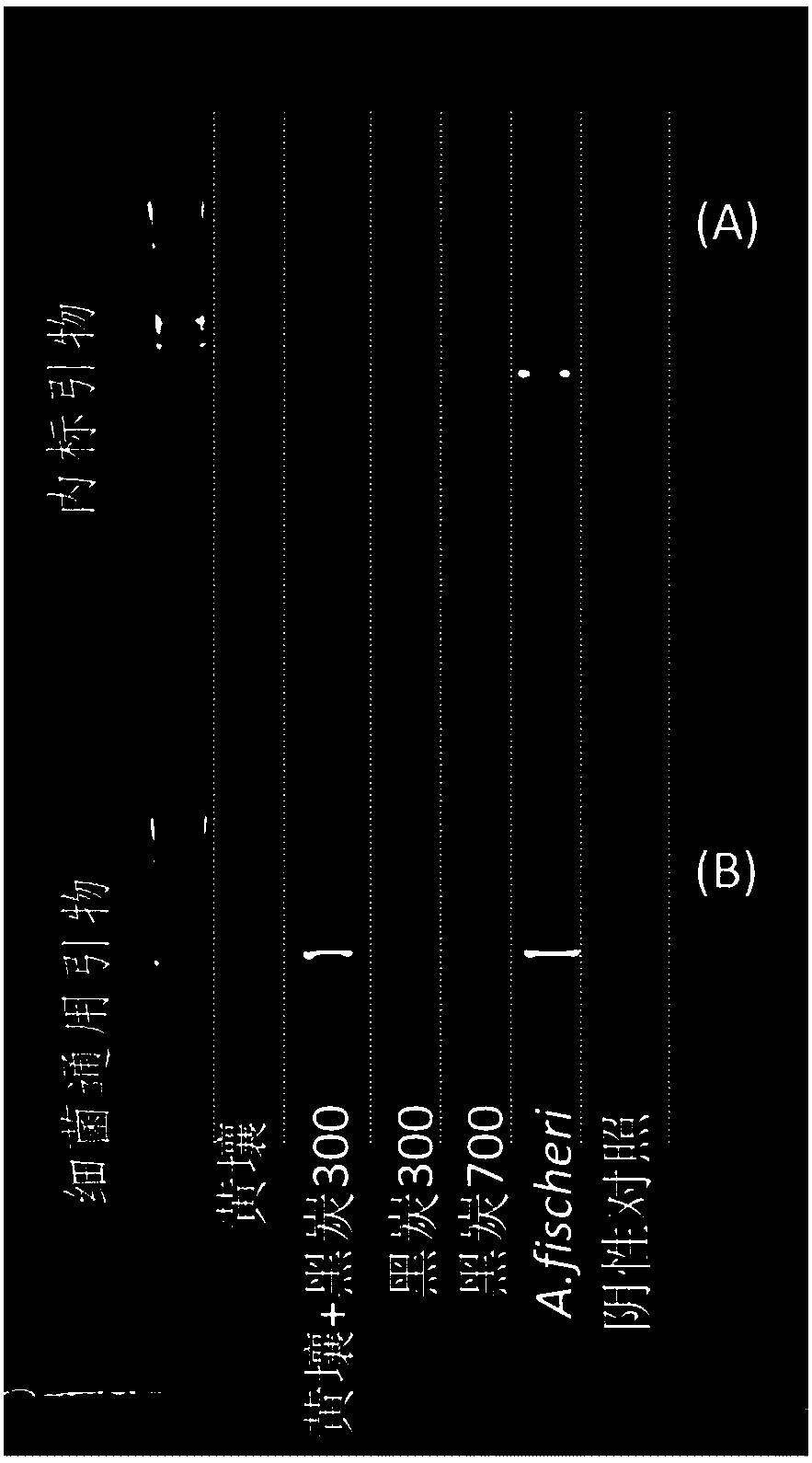
[0049] A), purchase A.fischeri ATCC 7744T bacterial strain, cultivate at room temperature (25 ℃) on the tryptone soybean agar medium that concentration is 3%NaCl, use strong The DNA was extracted by the DNA extraction kit, and its concentration was measured by Qubit 3.0, which was used as an internal standard for determining the DNA extraction rate of the soil sample; it was used for future use.
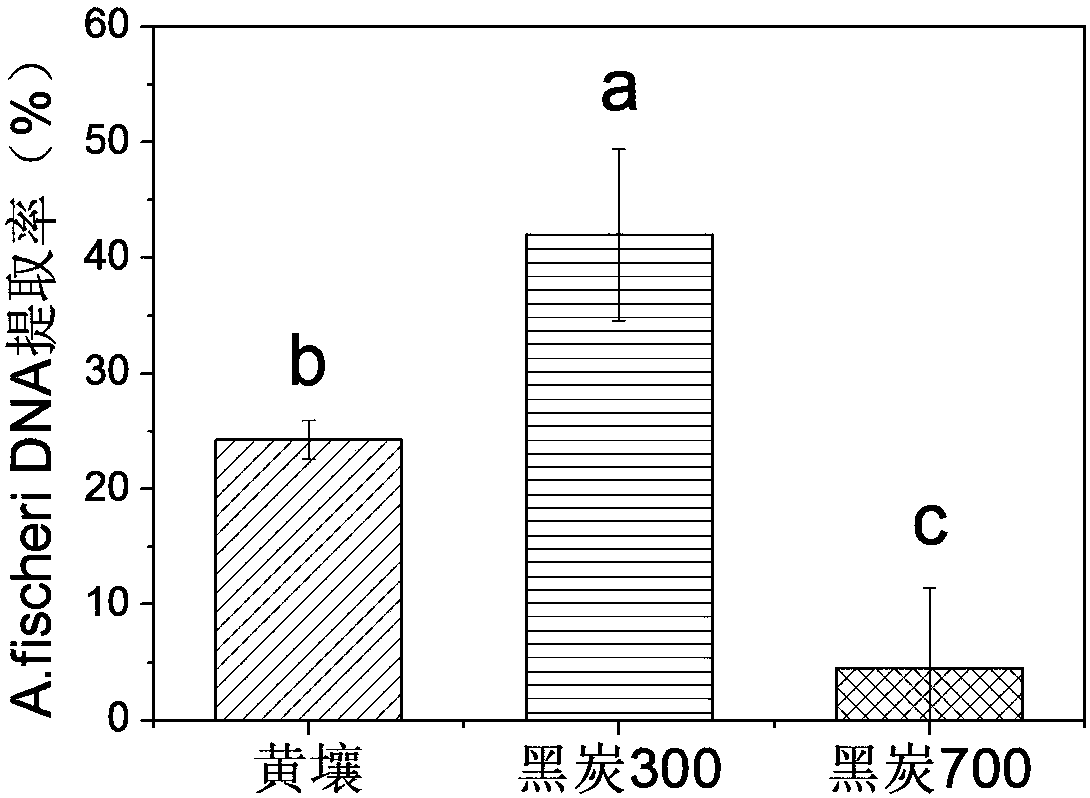
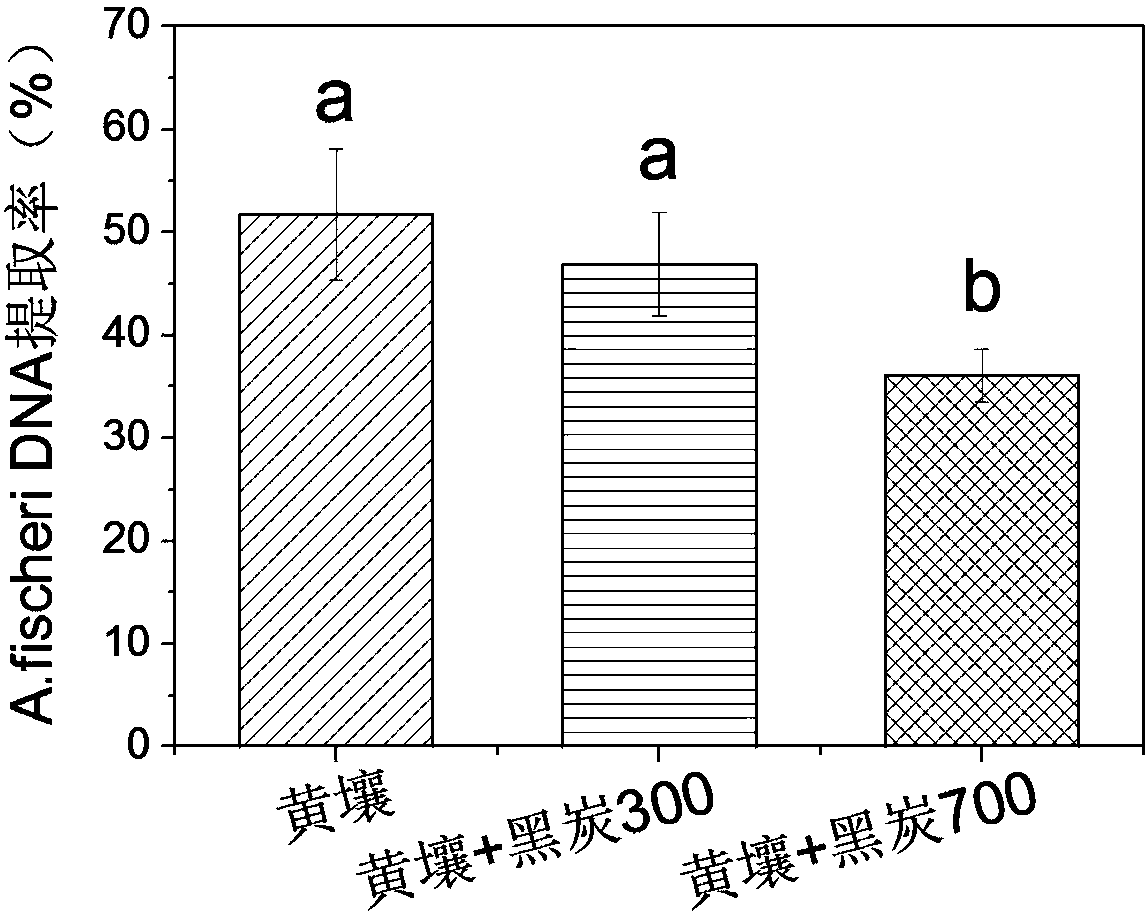
[0050] B) A mineral soil (yellow soil), a black carbon obtained by pyrolysis at 300°C (black carbon 300) and a black carbon obtained by pyrolysis at 700°C (black carbon 700) were used as test materials. Among them, Heitan 300 is rich in aliphatic carbon structure, while Heitan 700 is rich in aromatic carbon structure. The carbon structure of the two is very different, which can be regarded as two different types of soil organic matter. At the same ti...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap