Gentisic acid dioxygenase as well as coding gene and application thereof
A technology of dioxygenase and dioxygenase protein is applied in the application field of gentisate dioxygenase and its encoding gene, which can solve the problems that restrict environmental behavior and ecological safety of dicamba, metabolic pathways and molecular mechanisms. Clear and other problems, to achieve the effect of huge application potential and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1. Cloning of gentisate dioxygenase gene dsmD
[0030] 1.1 Search for gentisate dioxygenase gene
[0031] 1.1.1 Substrate spectrum experiment of Rhizorhabdus dicambivorans Ndbn-20, an efficient dicamba-degrading bacterium
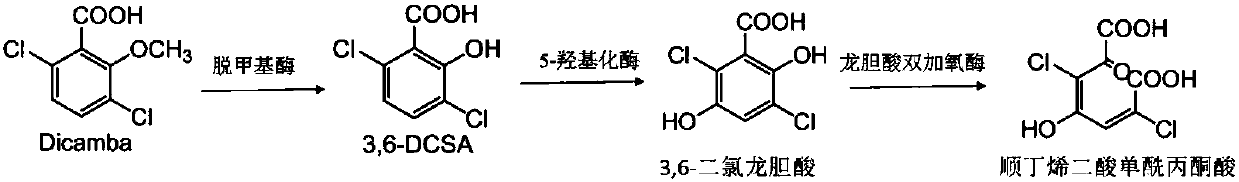
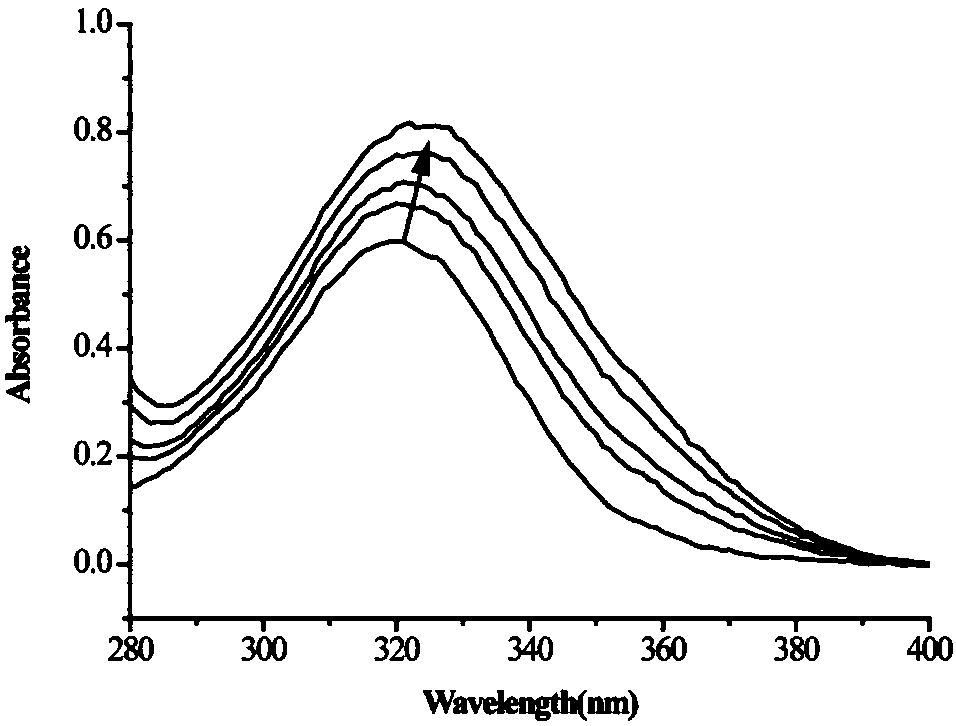
[0032] The research material of this experiment is Rhizorhabdus dicambivorans Ndbn-20, a high-efficiency dicamba-degrading bacterium isolated by members of our laboratory. The date is November 5, 2014. The substrate spectrum experiment of Ndbn-20 is to add appropriate concentration of 3,6-DCSA analogue to the inorganic salt medium, regularly sample and use the method of ultraviolet scanning to detect the degradation status, the degradation status is shown in Table 1. Through the substrate spectrum experiment, it was found that Ndbn-20 can degrade gentisic acid but not catechol, so it is speculated that the ring-opening substrate of 3,6-DCSA degradation reaction may be gentisic acid or gentisic acid analogue3 ,6-Dichlorogentisic acid.
...
Embodiment 2
[0044] Example 2 High expression of gentisate dioxygenase gene in BL21 (pET-24b (+))
[0045] 2.1 PCR amplification of gentisate dioxygenase gene
[0046] Forward primer: 5'-CGGGAATTC CATATG ACTGCTACGTCGATCAAACAC-3' (SEQ ID NO.3) and reverse primer: 5'-CCG CTCGAG Using CGCGCTACGCCAAAGGCCCAG-3' (SEQ ID NO.4) as a primer, a gentisate dioxygenase gene fragment was amplified from Ndbn-20 genomic DNA by PCR.
[0047] Amplification system:
[0048]
[0049] PCR amplification program:
[0050] a. Denaturation at 98°C for 3 minutes;
[0051] b. Denaturation at 98°C for 0.5min, annealing at 58°C for 0.5min, extension at 72°C for 1.0min, and 30 cycles;
[0052] c. Extend at 72°C for 10 minutes and cool to room temperature.
[0053] 2.2 Double digestion of PCR product and plasmid, product purification and enzyme connection
[0054] The PCR product was purified using a gel purification recovery kit. For specific methods, refer to the kit instructions. Purified PCR products an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com