Serine hydroxymethyltransferase gene promoter region methylation degree detection kit and method
A technology of serine hydroxymethyl group and gene promoter region is applied in the field of detection kits for the methylation degree of serine hydroxymethyltransferase gene promoter region, which can solve the problems of less research on the regulation mechanism of homocysteine and the like, Achieve the effect of low detection cost, strong specificity, and ensure accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
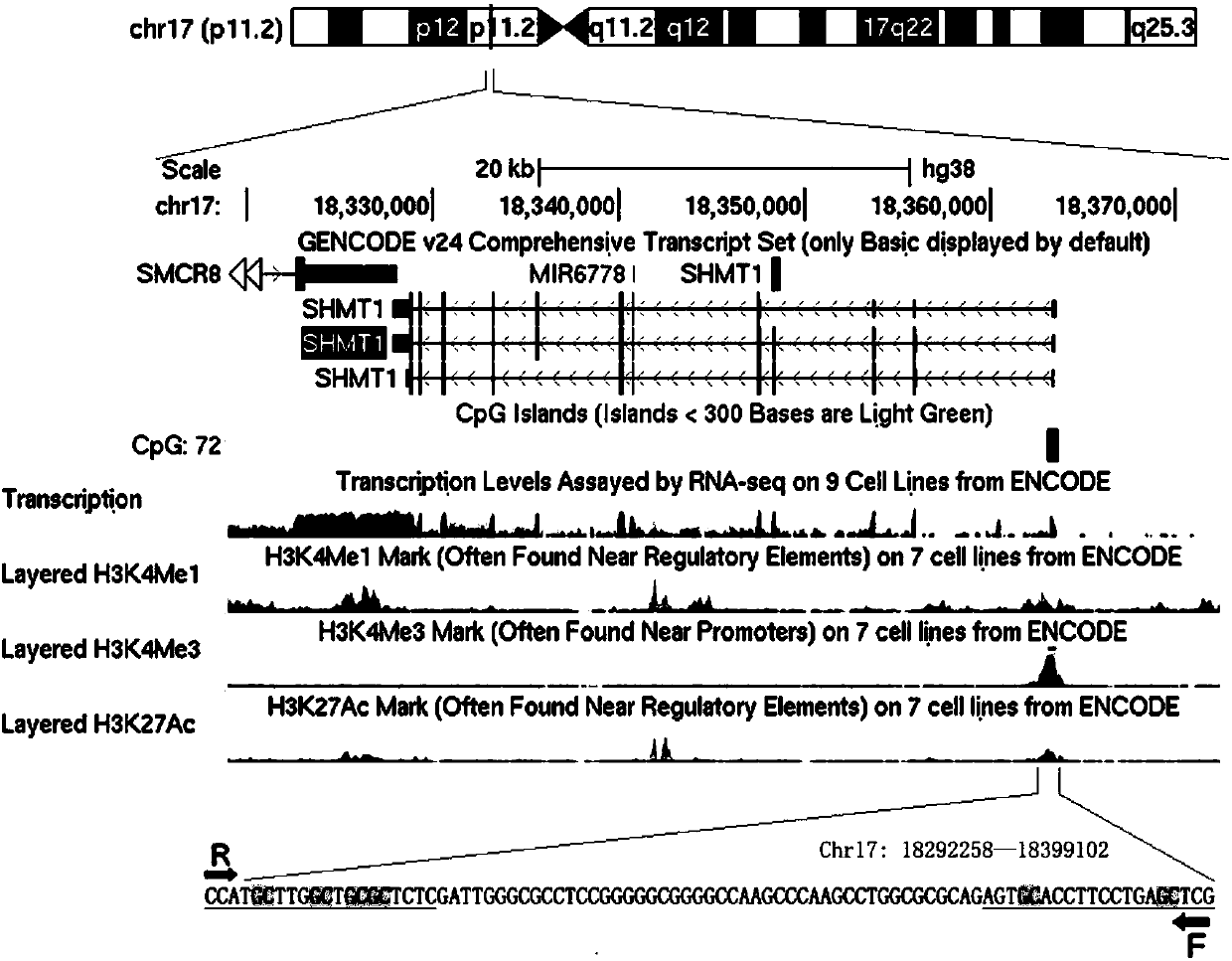
[0054] A detection kit for the degree of methylation in the promoter region of the serine hydroxymethyltransferase gene, the detection kit includes a DNA extract, a fluorescent quantitative PCR reaction solution, a positive control, a negative control, and a plurality of sealed tubes with caps. Fluorescent quantitative PCR reaction solution includes the PCR reaction solution as 10×PCR buffer 2μL, 25mmol / L MgCl 2 1.5μL, 2.5mmol / L d NTPs 1.5μL, 10μM Primer F 0.25μL, 10μM Primer R 0.25μL, 5U / μL hot-start Taq DNase 0.2μL, DNA template 1μL, 2×SYBR Green I 5μL, sterile double-distilled Make up to 20 μL with water; the nucleotide sequences of Primer F and Primer R are as follows:
[0055] Primer F: 5'-CGAGTTTAGGAAGGTGTATT-3' (SEQ ID NO.1) and
[0056] Primer R: 5'-CCATACTTAACTACGCTCTC-3' (SEQ ID NO.2);
[0057] The positive control is a complete DNA methylation sample obtained after the genome of a normal human sample is completely methylated; the negative control is sterile double...
Embodiment 2
[0059] A method for determining the degree of methylation in the promoter region of a serine hydroxymethyltransferase gene, comprising:
[0060] DNA extraction steps: use Lab-Aid 820 automatic nucleic acid extractor to extract whole blood genomic DNA and use NanoDrop2000 ultra-micro spectrophotometer to detect the concentration of DNA;
[0061] Using a methylation kit to perform bisulfite conversion on whole blood genomic DNA Step: Using the EZ DNAMethylation-Gold kit to react the above-mentioned extracted whole blood genomic DNA with bisulfite will make the unmethylated DNA in the single strand Cytosine is converted to thymine, and then converted DNA is obtained through steps such as loading, washing, and elution;
[0062] DNA methylation level determination steps: the above-mentioned converted DNA is used as sample DNA and added to the reaction solution containing fluorescent quantitative PCR for PCR amplification reaction, so that the gene sequence is multiplied. Each sampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com