Biosensors comprising allosteric transcription factor regulatory systems, kits and their use in small molecule detection
A technology of biosensors and transcription factors, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, and analysis through chemical reactions of materials, which can solve the problems of difficulty in lowering the detection limit and poor detection stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0155] Example 1: Uric acid biosensor
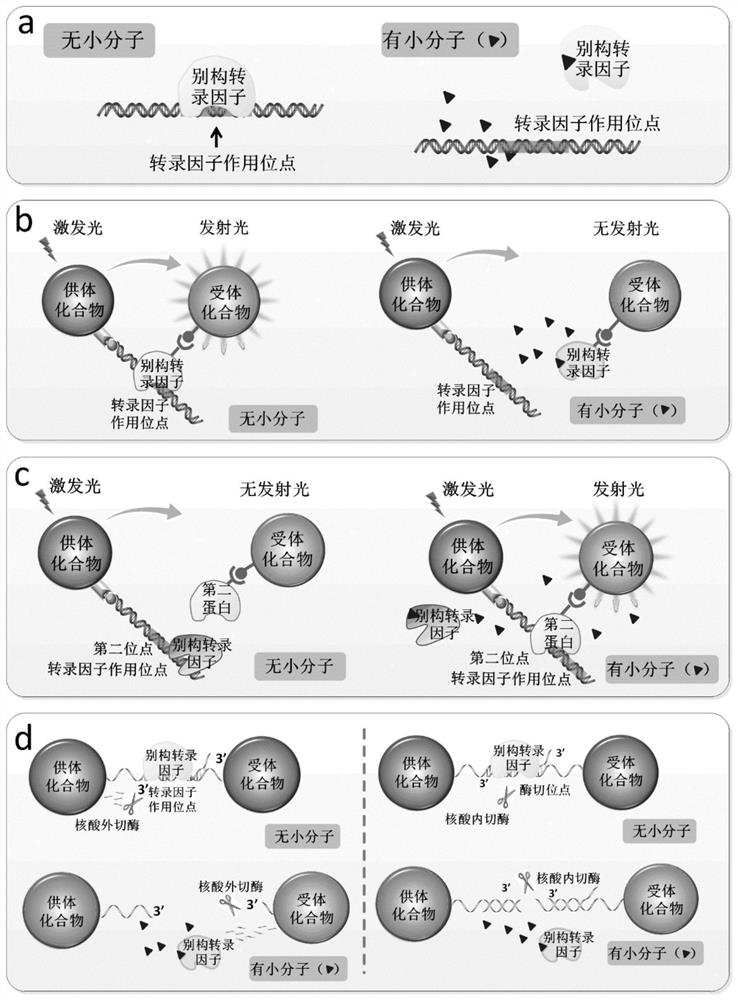
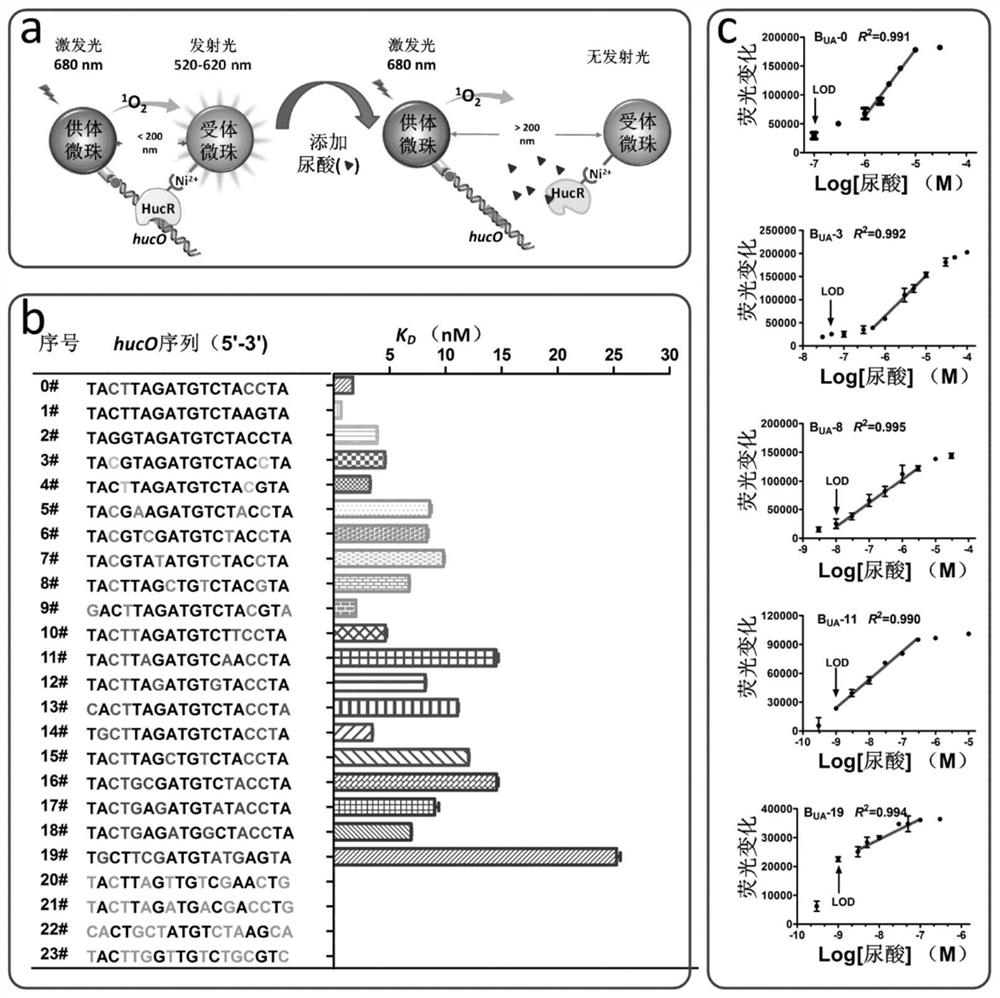
[0156] In this example, the allosteric transcription factor HucR and the DNA fragment hucO interacting with it are used as the recognition element, and the ALPHA technology microbeads are used as the transducer element to construct a biosensor platform capable of sensing uric acid, and use a mathematical model to analyze the sensitivity of the biosensor It provides a general, systematic and effective strategy to improve the sensitivity and detection range of biosensors.
[0157] The HucR protein is a repressor protein derived from Deinococcus radiodurans. In the absence of uric acid (UA), HucR can bind to its operator site hucO, preventing the transcription of downstream target genes. In the presence of uric acid, uric acid specifically binds to HucR, releasing HucR from hucO and turning on the transcription of target genes. Therefore, the concentration of uric acid in the sample can be determined from the degree of binding of HucR to ...
Embodiment 2
[0273] Embodiment 2: oxytetracycline biosensor
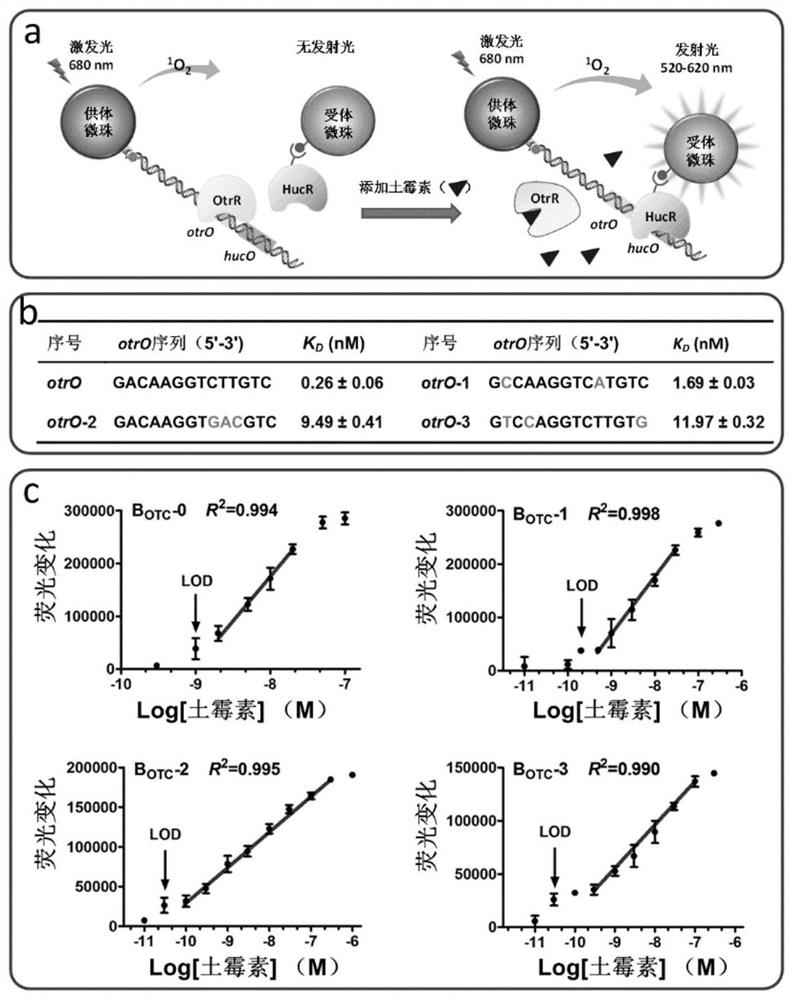
[0274] In this example, the allosteric transcription factor OtrR and the DNA fragment containing its action site otrO are used as the recognition element, the donor beads and acceptor beads of ALPHA technology are used as the first transduction element, and HucR and its interaction site hucO are used as the second transducing element. The second transducer element is used to construct a biosensor platform capable of sensing oxytetracycline. The DNA fragment has both otrO and hucO fragments ( image 3 a).
[0275] OtrR is a repressor protein derived from Streptomyces chamotis. In the natural state, in the absence of oxytetracycline, OtrR can bind to its operator site otrO, preventing the transcription of downstream target genes. In the presence of oxytetracycline, oxytetracycline specifically binds OtrR, releasing OtrR from otrO and turning on the transcription of target genes. Therefore, the concentration of oxytetracycline ...
Embodiment 3
[0305] Embodiment 3: prunamycin biosensor
[0306]In this example, two kinds of prunamycin biosensors using nuclease as the second transduction element were constructed. The recognition elements of the sensor are all allosteric transcription factors Pip and ptr sites, and the first transducer elements are all gold nanoparticle pairs. In an embodiment utilizing an exonuclease, exonuclease III and a DNA fragment with a 3' overhang are used as the second transducing element. In embodiments utilizing an endonuclease, a HindIII endonuclease and a HindIII restriction site are used as the second transducing element.
[0307] Pip is a repressor protein derived from Streptomyces coelicolor. In the natural state, in the absence of prunamycin, Pip can bind to its operator site ptr, preventing the transcription of downstream target genes. Prinamycin specifically binds Pip when it is present, releasing Pip from ptr and turning on the transcription of the target gene. Therefore, the con...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com