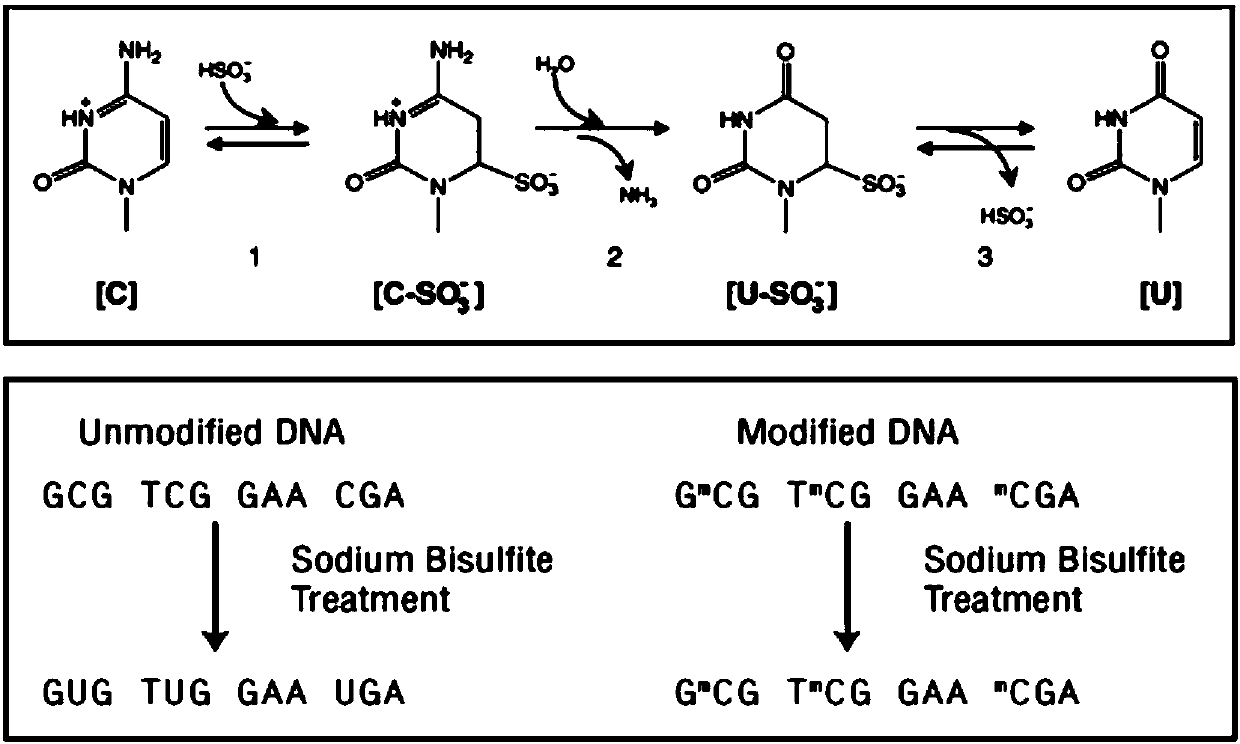
Nucleic acid combination and kit for detecting septin9 gene methylation
A methylation and kit technology, applied in the field of molecular biology, can solve the problems of decreased sample detection rate, low colorectal cancer specificity, low screening penetration rate, etc., to improve sensitivity and specificity, prevent false Positive results, the effect of shortening the test time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] This embodiment provides a kit for detecting the methylation of the septin9 gene, which includes: a nucleic acid combination for detecting the methylation of the septin9 gene, a Blocker sequence, a pair of primers for the β-actin internal reference gene, a probe for the β-actin internal reference gene, sub- Sulfate conversion reagent set, 2×PCR Mix and taq enzyme.
[0058] Wherein, the nucleic acid combination includes: a pair of primers for detecting the methylation of the septin9 gene and a probe for detecting the methylation of the septin9 gene.
[0059] The primer pair for detecting the methylation of the septin9 gene includes: an upstream primer whose base sequence is shown in SEQ ID NO.11 and a downstream primer shown in SEQ ID NO.17.
[0060] The base sequence of the septin9 gene methylation detection probe is shown in SEQ ID NO.19.
[0061] Among them, the 5' end of the septin9 gene methylation detection probe is labeled with a fluorescent reporter group FAM, a...
experiment example 2
[0100] The septin9 methylation detection site in the present invention includes 6 methylation sites located in the third CpG island of the septin9 promoter region. In this experimental example, the forward sequence of the target sequence where the methylation site is located is used as a template to design methylation-specific reference primers and probe sequences, as shown in Table 1.
[0101] At the same time, using the β-actin gene as an internal reference, β-actin control primers and probes were designed, as shown in Table 1.
[0102] Table 1 Primer and Probe Sequences
[0103]
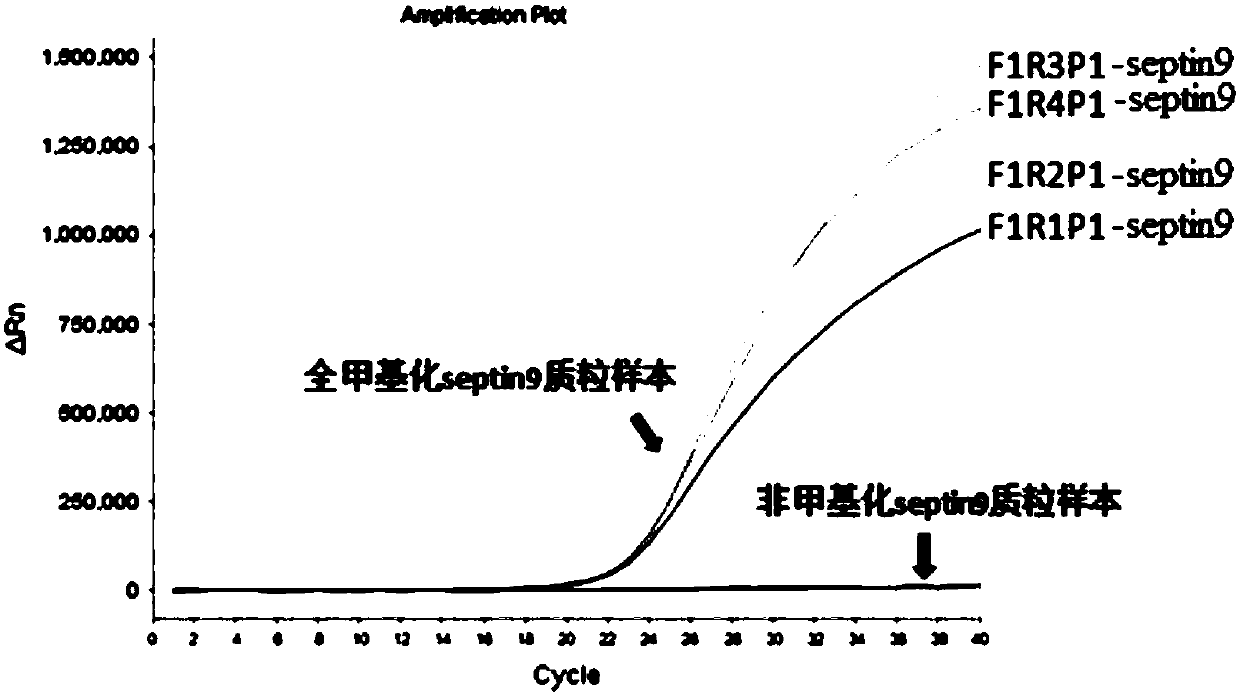
[0104] The nucleic acid combination (primer: SEQ ID NO.11 (septin9-F1), SEQ ID NO.17 (septin9-R3), probe: SEQ ID NO. ID NO.19 (septin9-P1)) and β-actin internal reference gene primer pair and probe (primers: SEQ ID NO.1 (β-actin-F1), SEQID NO.6 (β-actin-R3), probe Needle: SEQ ID NO.7 (β-actin-P1)) for amplification efficiency verification, as follows.
[0105] Reagents: 2×PCR Mix, taq DNA poly...
experiment example 3
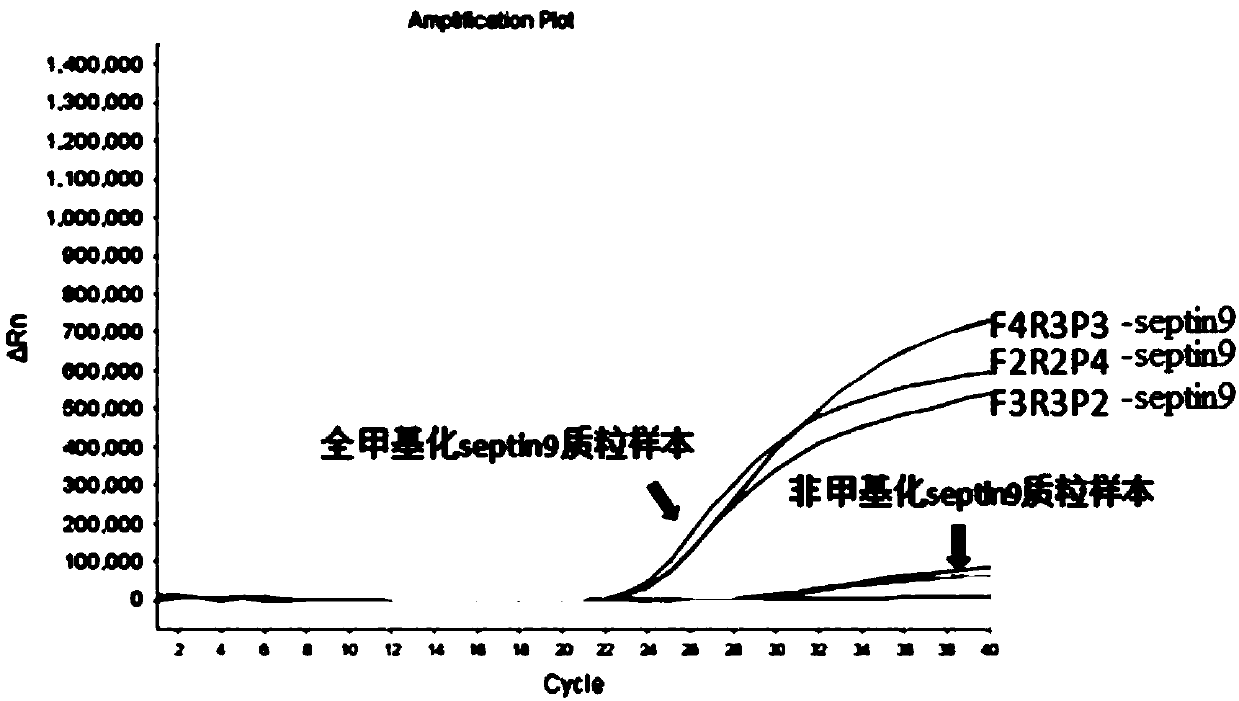
[0117] Dilute the methylated septin9 synthesis plasmid to 10 -2 ng / μl, 10 -3 ng / μl, 10 -4 ng / μl, 10 -5 ng / μl, 10 -6 5 concentration gradients of ng / μl were used as DNA samples, and the copy numbers corresponding to each concentration were shown in Table 3, and the kit provided in Example 1 was used for qPCR amplification to verify the sensitivity of the kit provided in Example 1 . The result is as image 3 shown.
[0118] Table 3 Correspondence between copy number and plasmid concentration
[0119] Concentration (ng / μl)
[0120] according to Figure 6 The results showed that with the decrease of the concentration, the fluorescence value and ct value of the sample showed a downward trend. The sample concentration is 10 -6 ng / μl (10 2 copy), the amplification curve was a smooth S-shape. Continue to reduce the sample concentration, the amplification curve is not S-shaped (results not shown), which affects the result judgment. Therefore determine that the det...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com