Primer and probe group and kit for detecting hemolytic streptococcus through RAA fluorescence method
A hemolytic streptococcus, primer probe technology, applied in the direction of DNA / RNA fragments, microorganisms, recombinant DNA technology, etc., can solve the problems of high false positives, complex design, etc., to achieve high specificity detection, rapid detection, large The effect of applying the foreground
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
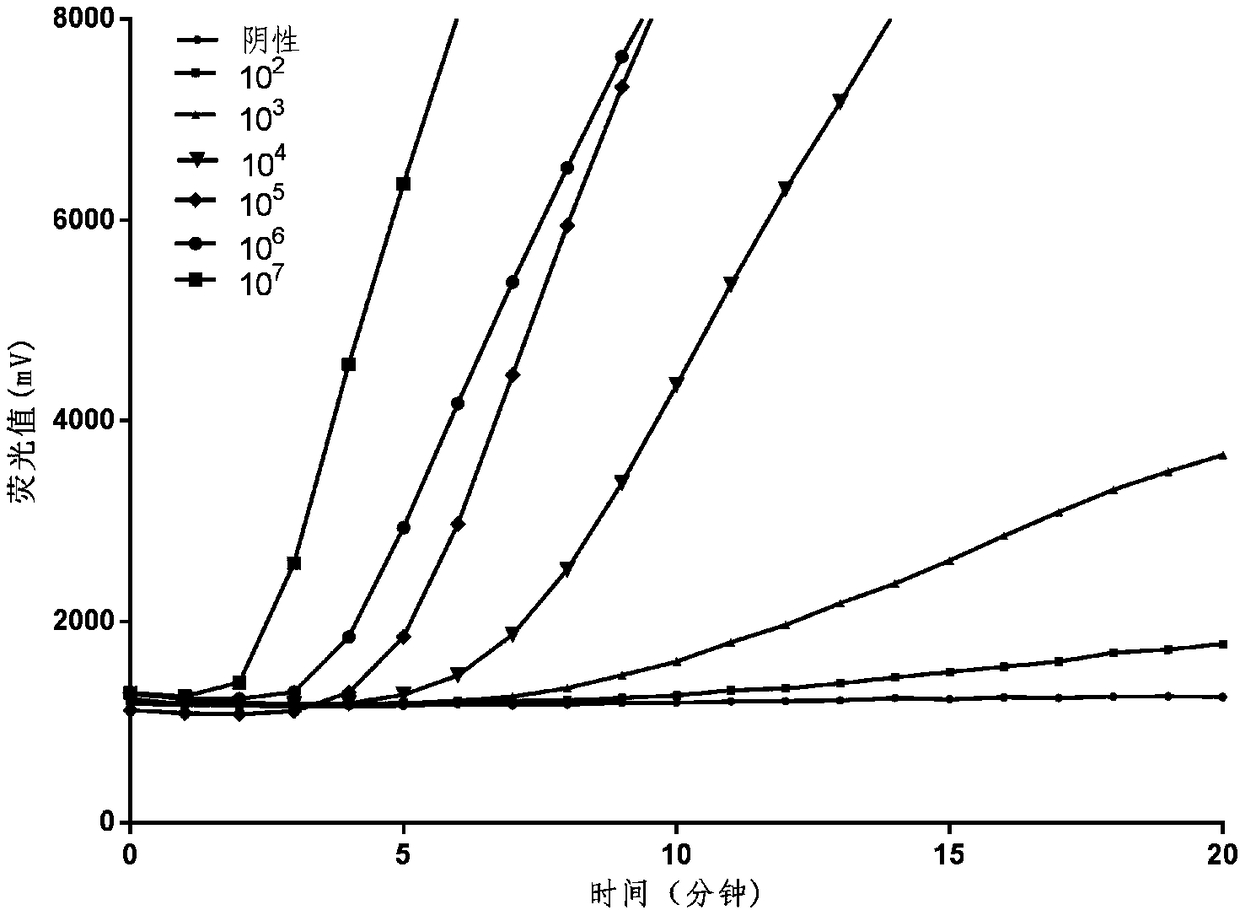
[0054] Select the specific conserved gene of hemolytic streptococcus as the target detection gene, and obtain the hemolytic streptococcal protease A gene through the National Center for Biotechnology Information (NCBI) (http: / / www.ncbi.nlm.nih.gov). Sequence, and perform multiple sequence alignment, and select a conservative sequence, the sequence is shown in SEQ ID NO.6 in the sequence listing:
[0055] TGACAACAACAGTAGCAGCAGATGAGCTAACCACAACGAGTGAACCAACAATCACGAATCACACTCAACAACAAGCGCAACATCTCACCAATACAGAGTTGAGCTCAGCTGAATCAAAACCTCAAGACACATCACAAATCACTCTCAAGACAAATCGTGAAAAAGAGCAACCACAAGGTCTAGTCTCTGAGCCAACCACAACTGAGCTAGCTGACACAGATGCAGCACCAATGGCTAATACAGGTCCTGATGCGACTCAAAAAAGCGCTTCTTTACCGCCAGTCAATACAGATGTTCACGATTGGGTAAAAACCAAAGGAGCTTGGGACAAGGGATACAAAGGACAAGGCAAGGTTGTCG;
[0056] According to the above sequence, Sangon Bioengineering (Shanghai) Co., Ltd. was commissioned to synthesize a DNA plasmid with a size of 360bp;
[0057] According to the design principles of RAA technical primers ...
Embodiment 2
[0084] The primer probe is the same as in Example 1.
[0085] Table 4 is the composition of embodiment 2 kit
[0086]
[0087] Sample source and DNA extraction
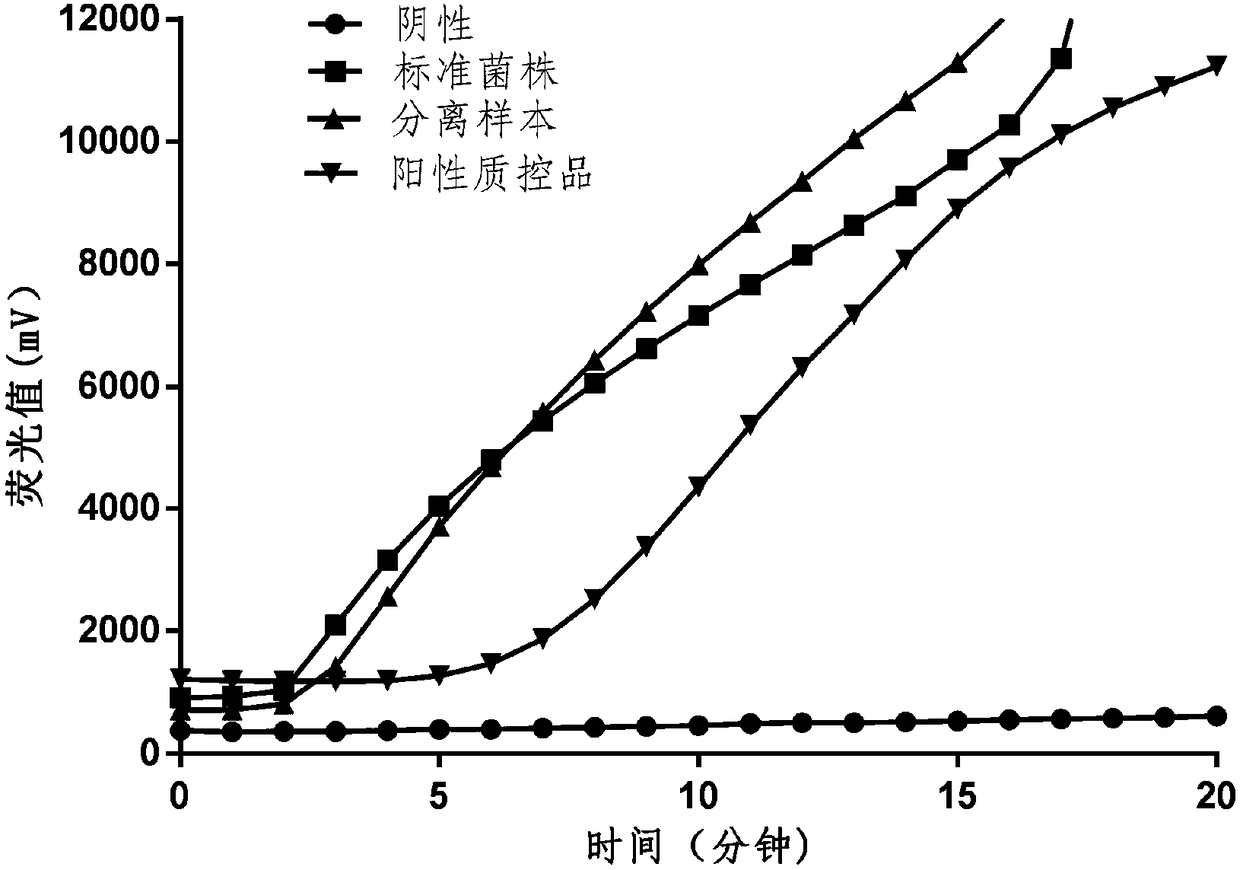
[0088] The samples were provided by Zhejiang Entry-Exit Inspection and Quarantine Bureau. They were DNA extracted from standard strains of hemolytic streptococci and samples of hemolytic streptococci isolated from food. The extracted DNA was stored at -80°C for later use.
[0089] Take 4 reaction tubes and operate as follows respectively, absorb 27 μL of reaction buffer, add 2 μL of the mixture of probe and primer, and mix thoroughly; add 49 μL of the mixed buffer into the RAA basic fluorescent universal reaction reagent tube, Make the lyophilized powder fully dissolve and mix;
[0090] Add 1 μL negative quality control substance, 1 μL extracted hemolytic streptococcus standard strain DNA, 1 μL extracted hemolytic streptococcus sample DNA, and 1 μL positive quality control substance to the 4 prepared tubes as tem...
Embodiment 3
[0096] The sequences of the primer probe and the positive quality control product are the same as those in Example 1.
[0097] Table 5 is the composition of embodiment 3 kit
[0098]
[0099]
[0100] Sample source and DNA extraction:
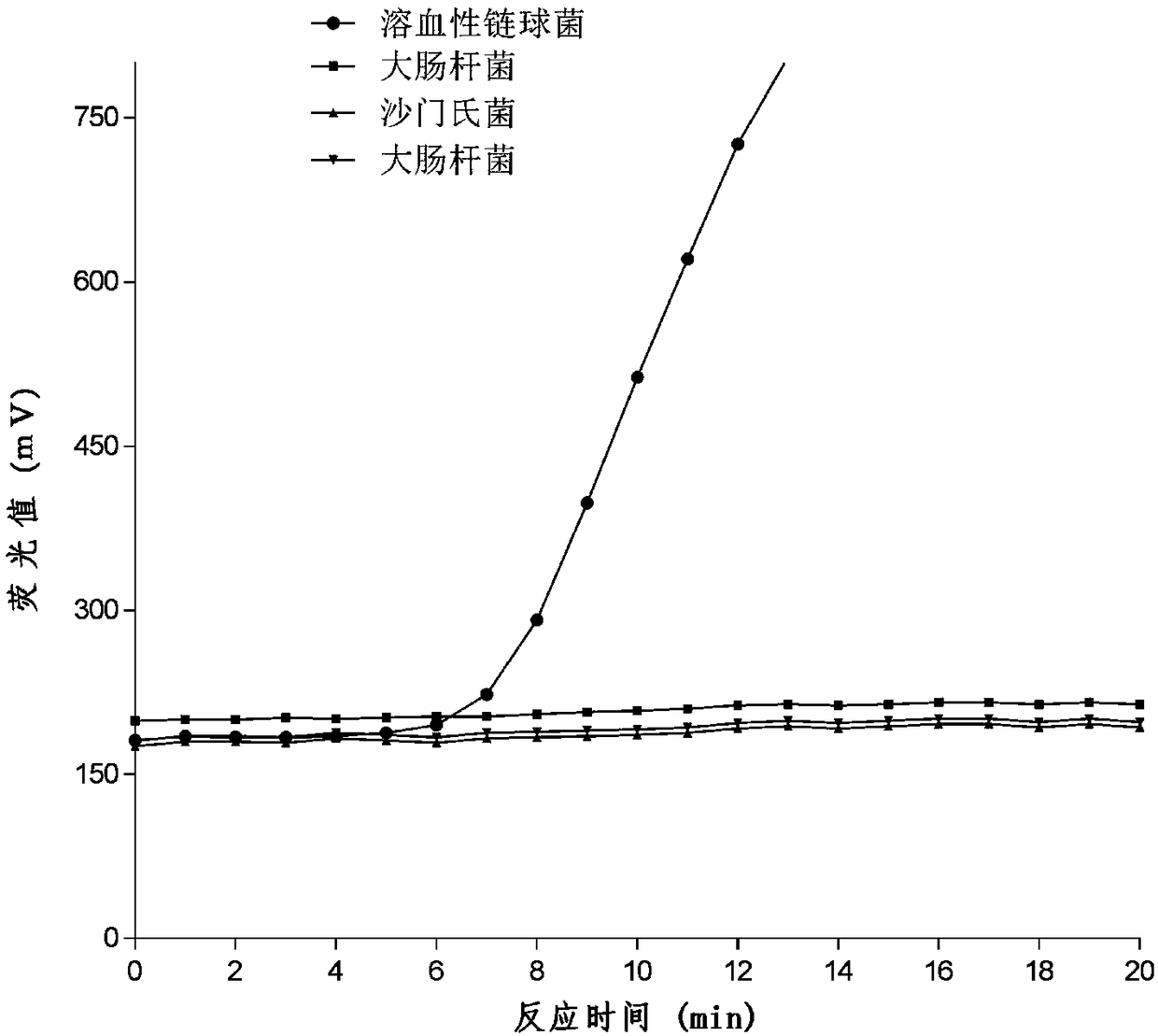
[0101] The DNA of hemolytic streptococcus samples was provided by Zhejiang Entry-Exit Inspection and Quarantine Bureau, and the samples of Salmonella, Staphylococcus aureus and Escherichia coli were provided by Zhejiang International Travel Health Center. DNA was extracted by heating at 90°C for 10 minutes, and the extracted DNA was stored at -80°C spare,
[0102] Take 5 reaction tubes and operate as follows respectively, absorb 27 μL of reaction buffer, add 2 μL of the mixture of probe and primer, and mix thoroughly; add 49 μL of the mixed buffer into the RAA basic fluorescent universal reaction reagent tube, Make the lyophilized powder fully dissolve and mix;
[0103] Add 1 μL of negative quality control, 1 μL of extracted hemolytic ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com