Method for sorting and screening nano antibody through flow cytometry
A nanobody, sorting and screening technology, used in chemical instruments and methods, botanical equipment and methods, biochemical equipment and methods, etc. The effect of avoiding phage contamination, less workload, and shorter screening cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Using Flow Cytometry to Screen Anti-Pseudomonas Exotoxin A Nanobodies
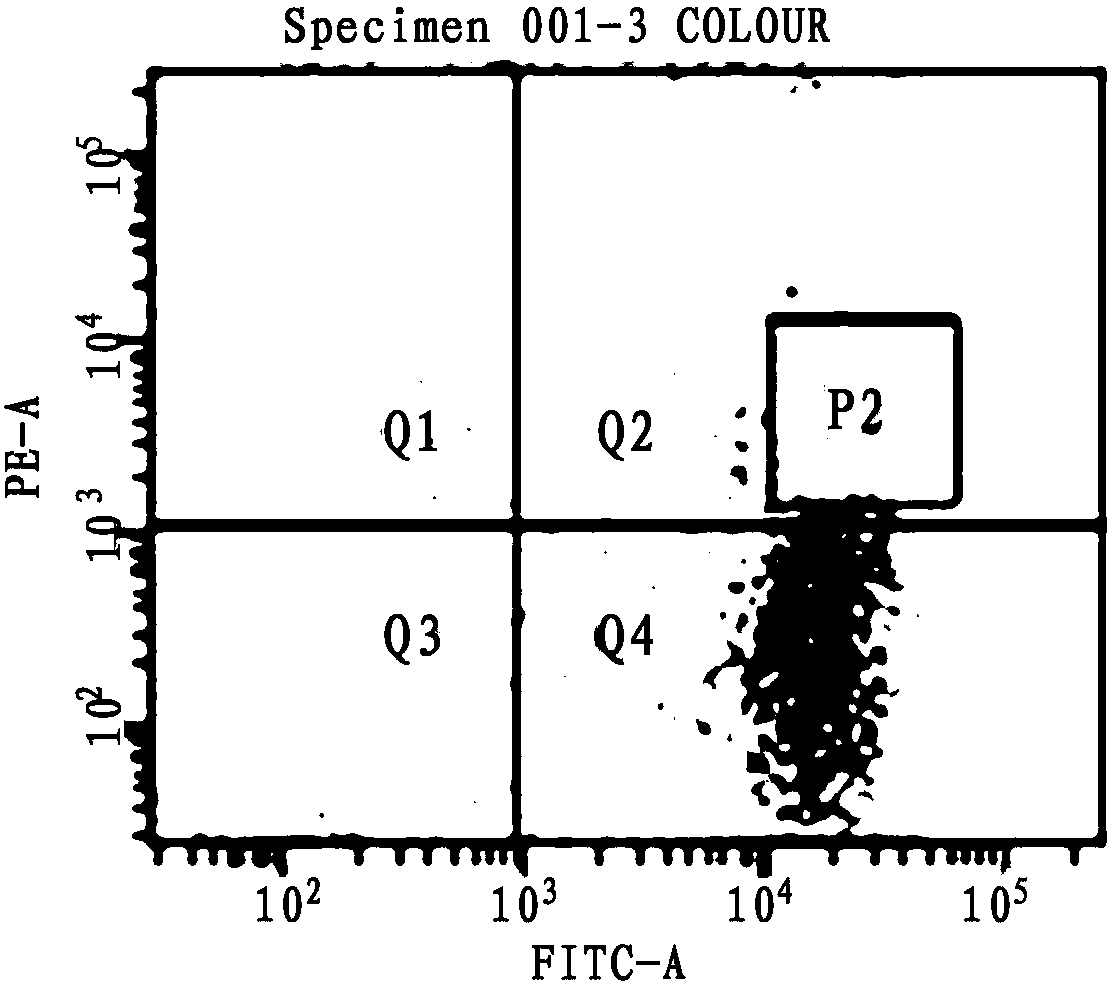
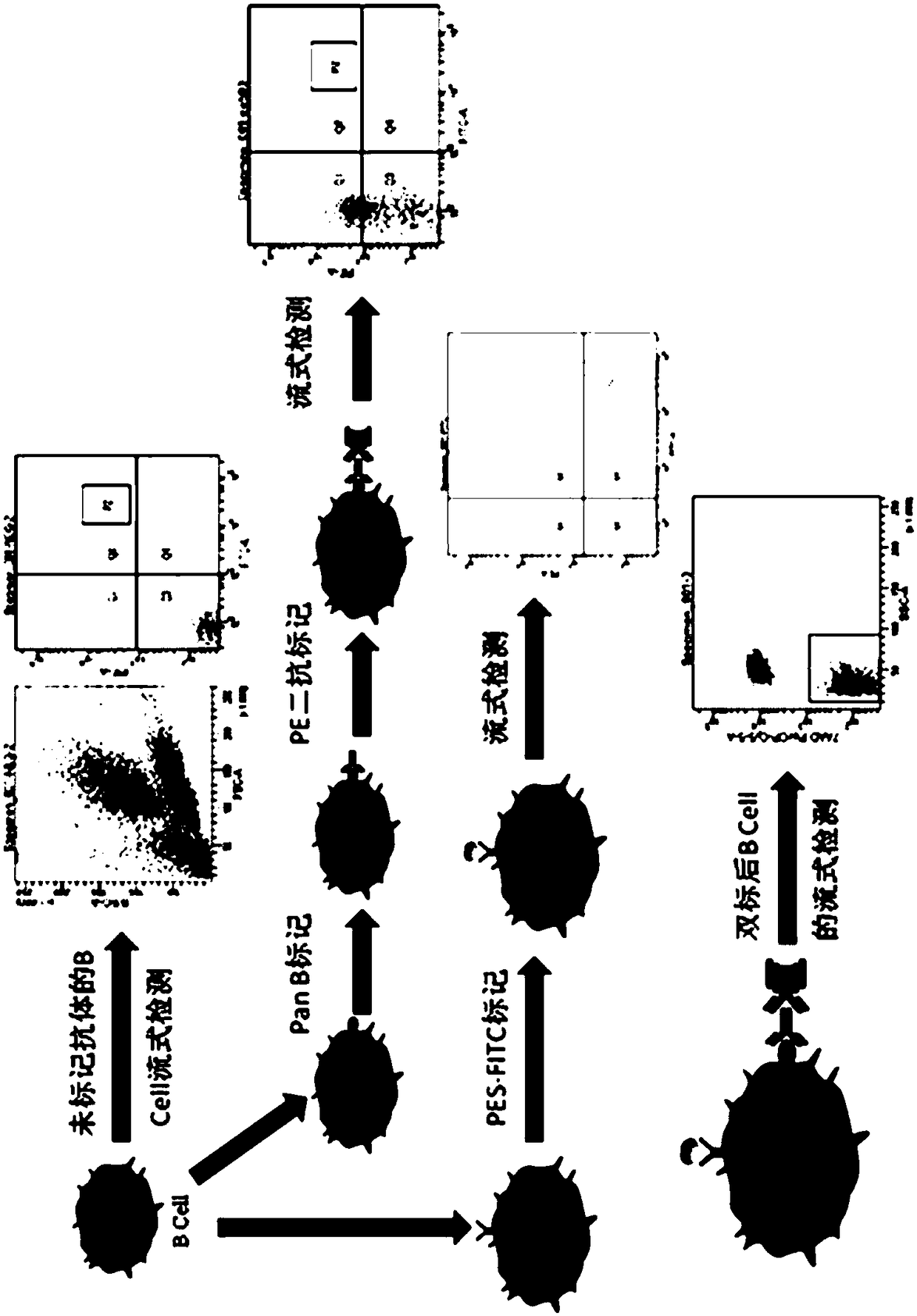
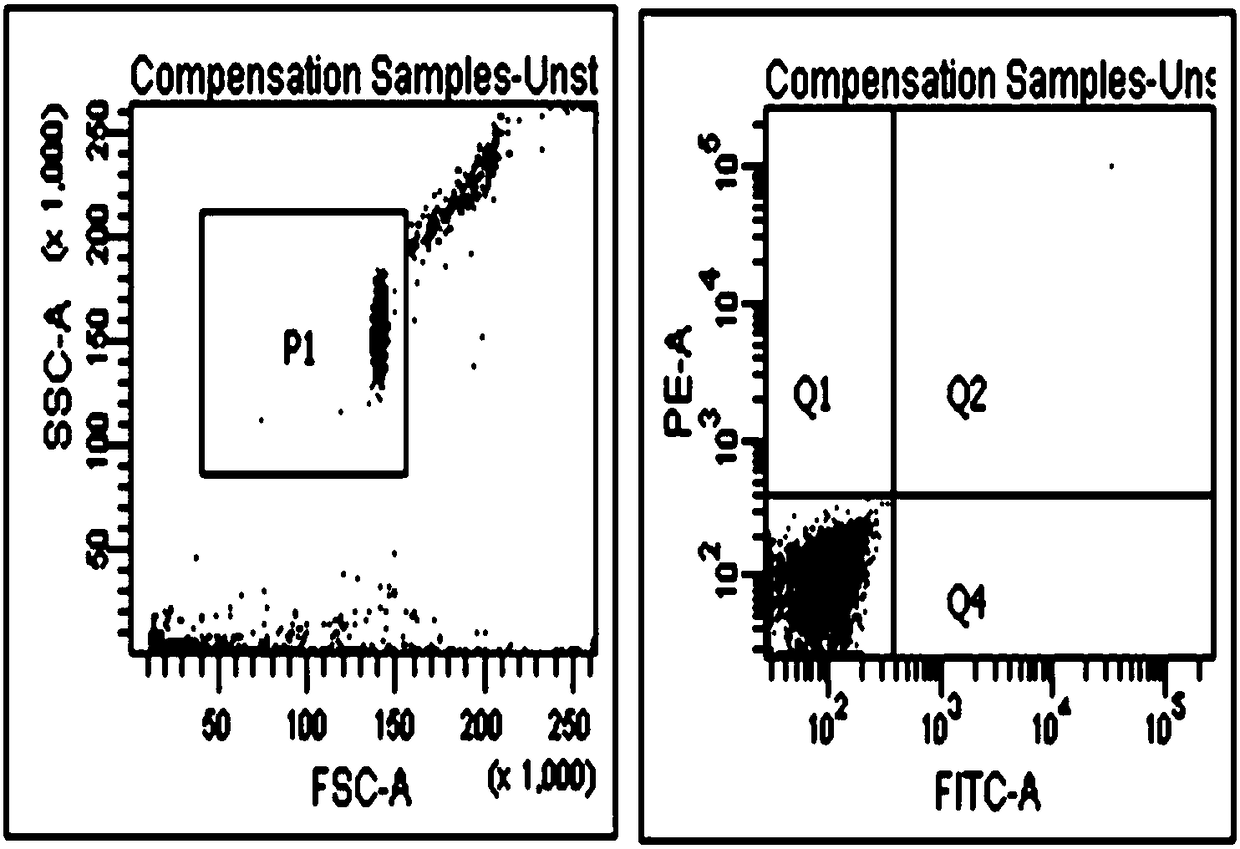
[0052] figure 1 A preferred embodiment of the method disclosed in the present invention is given in figure 1 Firstly, B cells with unlabeled antibody were used for flow cytometry detection, and this was used as a negative control; at the same time, Anti-llama pan B cell monoclonalantibody was used as the primary antibody, Rat Anti-Mouse IgM-PE was used as the secondary antibody to label B cells, and FITC was used to label B cells Pseudomonas aeruginosa exotoxin A; establish the FSC-H / FSC-W experimental plate, and finally obtain PE and FITC double positive samples.
[0053] 1.1 Antigen-immunized alpaca: A healthy adult alpaca was selected, and the peripheral blood of the alpaca was collected as a negative control not immunized with antigen, and stored in a -80°C ultra-low temperature refrigerator. Mix the recombinant Pseudomonas aeruginosa exotoxin A as antigen with Freund's adjuvant at a...
Embodiment 2
[0099] Example 2. Analysis of affinity activity between nanobody and antigen Pseudomonas aeruginosa exotoxin A
[0100] 2.1 Chip antigen coupling: Prepare the antigen with different pH sodium acetate buffer (pH 5.5, pH 5.0, pH 4.5, pH 4.0) to prepare 20 μg / mL working solution, and prepare 50 mM NaOH regeneration solution at the same time, use Biacore The template method in the instrument of the T100 protein interaction analysis system analyzes the electrostatic binding between antigens at different pH conditions and the surface of the chip (GE Company). Use a neutral pH system and adjust the antigen concentration as needed for coupling conditions. The chip was coupled according to the template method that comes with the instrument: select the blank coupling mode for channel 1, select the Target coupling mode for channel 2, and set the target to the designed theoretical coupling amount. The coupling process takes about 60 min.
[0101] 2.2 Exploration of analyte concentration...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com