Biomarker for forecasting esophageal squamous cell carcinoma prognosis
A technology of biomarkers and prognostic markers, applied in the application of biomarkers to predict the prognosis of esophageal squamous cell carcinoma, and in the field of biomarkers to predict the prognosis of esophageal squamous cell carcinoma, can solve the problems that did not take into account esophageal squamous cell carcinoma , to achieve the effect of increasing the degree of certainty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1 Screening of prognostic markers based on public big data
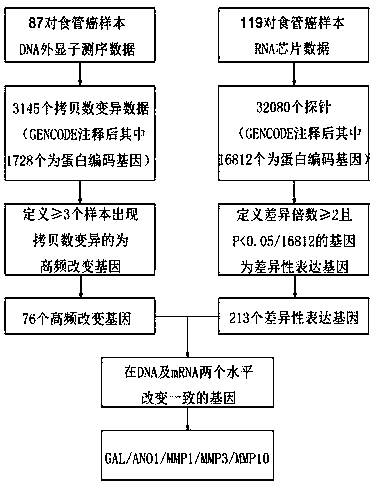
[0067] see figure 1 As shown, based on the exome sequencing data of 113 pairs of esophageal squamous cell carcinoma and paracancerous tissue DNA samples published earlier in the Chinese Han population (Gao YB, et al. Genetic landscape of esophageal squamouscell carcinoma. Nature genetics 2014;46: 1097 -1102.), the research analysis found that 87 pairs of samples contained a total of 3145 DNA copy number variation information. Using the GENCOD V19 gene annotation tool (http: / / www.gencodegenes.org / ) to confirm the copy number variation information of these genes, it was found that 1728 genes with protein coding function showed copy number variation, of which 76 genes were in Copy number variants (high frequency variants) found in three or more tissue samples.
[0068] Then we further retrieved the gene chip data information of the same cohort of patients with esophageal squamous cell carcinoma (GEO dat...
Embodiment 2
[0071] Example 2 Validation of Prognostic Markers
[0072] 1. Fabrication of Tissue Chips
[0073] 197 cases and 118 cases of esophageal squamous cell carcinoma patients and adjacent tissues were extracted from the Chinese Academy of Medical Sciences Cancer Hospital and the First Affiliated Hospital of Nanjing Medical University, respectively. There are complete clinical and pathological data and prognosis follow-up data. All tissue samples were firstly stained with hematoxylin-eosin after paraffin embedding to confirm that the paraffin tissue block was esophageal squamous cell carcinoma or paracancerous tissue, and then sliced to select a representative tumor area (cancer cells>80%) or Paracancerous area (0 for cancer cells). For each specimen, cancerous tissue or paracancerous tissue was selected twice, and samples from the two centers were placed on different tissue chips.
[0074] 2. Hematoxylin-eosin staining, including the following steps:
[0075] a. Place the tis...
Embodiment 3
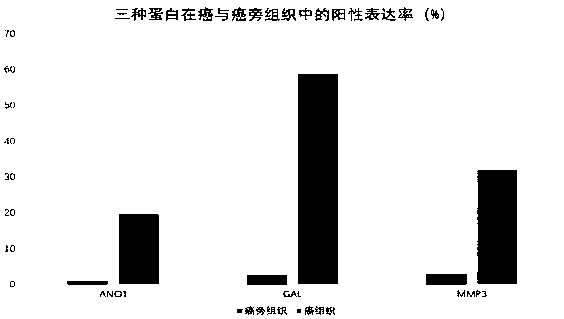
[0109] Example 3 Efficacy detection of prognostic markers - high expression of ANO1 promotes proliferation, apoptosis resistance, invasion and migration of esophageal squamous cell carcinoma cells
[0110] a. Cell culture
[0111]Human esophageal squamous cell carcinoma cell lines KYSE30, KYSE70, KYSE140, KYSE180, KYSE410, ECA109, TE-1 and normal esophageal epithelial cells HET-1a were purchased from Shanghai Jikai Biological Company. HET-1a cells were cultured in DMEM medium, other cancer cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum and 1% penicillin / streptomycin, the culture temperature was 37°C, and the carbon dioxide concentration in the culture environment was 5%, of which DMEM medium and RPMI-1640 medium were purchased from Gibco, USA.
[0112] b. RNA extraction and PCR detection
[0113] According to the requirements of the kit instructions, the total RNA extraction kit SuperfecTRI was used to extract RNA from the cells. The extracted tot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com