A method and kit for capturing nucleic acid-binding proteins
A protein protectant and protein technology, which is applied in the field of capturing nucleic acid binding proteins, can solve problems such as the inability to effectively obtain nonpolyA-RBPs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
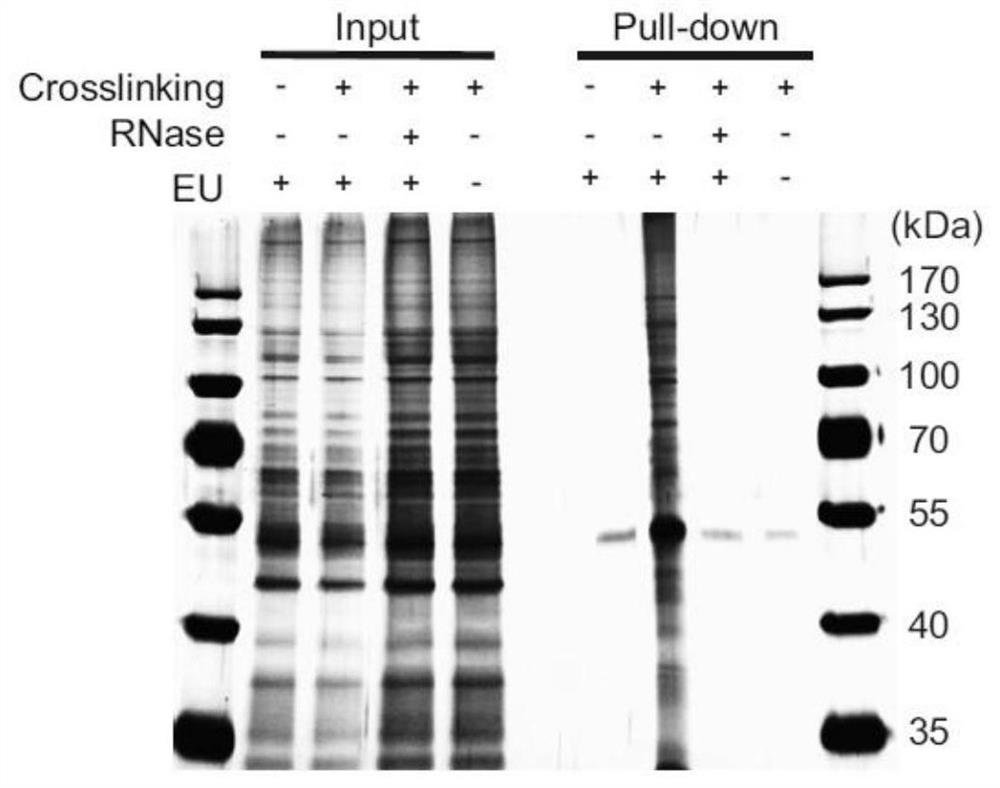
[0072] Example 1: Capture of RBP (RNA Binding Protein)
[0073] 1. Capturing RNA-protein complexes
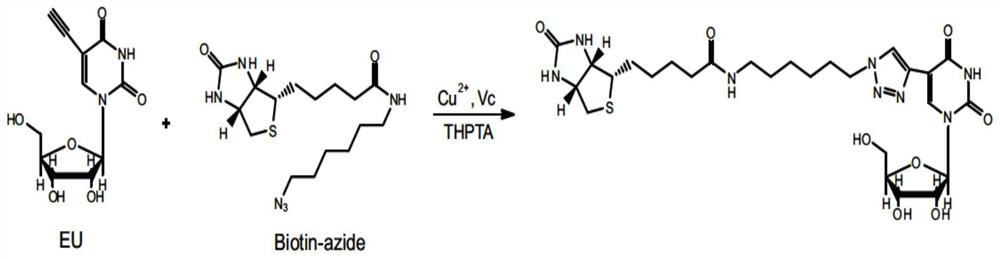
[0074] 1.1EU incorporated into RNA
[0075] HeLa (obtained from ATCC) cells were cultured in a petri dish with a diameter of 100 mm, and the culture medium was 10 mL of high-glucose medium containing 10% FBS at 37°C, 5% CO 2 cultivated under conditions. When the cells were cultured to a density of 80%, EU was added at a final concentration of 0.25 mM, and the cells were co-incubated with EU for 16 hours to fully incorporate EU into the newly synthesized RNA in the cells.
[0076] 1.2 Washing
[0077] Cells were washed twice with PBS buffer to remove residual medium.
[0078] 1.3 Photocrosslinking
[0079] Remove PBS, place the culture dish containing the cells of step (2) on ice, 254nm (0.4J / CM 2 ) UV light for 1 min to form a covalent bond between RBP and interacting RNA.
[0080] 1.4 Fixed cells
[0081] Remove the culture dish from the ultraviolet light, add 5 mL of ...
Embodiment 2
[0143] Embodiment 2: the dose optimization of protein protection agent
[0144] Detection of RBP by silver staining method: see the test results Figure 14 . (see Table 3 for the setting of test conditions, and refer to Example 1 for other steps.)
[0145] Table 3 Conditions of protein protectant
[0146] protein protectant THPTA concentration Aminoguanidine Concentration Condition 1 0mM 1mM Condition 2 0.3mM 1mM Condition 3 0.9mM 1mM Condition 4 1.5mM 1mM Condition 5 0.9mM 0.5mM Condition 6 0.9mM 1mM Condition 7 0.9mM 1.5mM Condition 8 0.9mM 0mM
[0147] Such as Figure 14 As shown, when THPTA is not added to the click reaction solution (condition 1), the protein bands can be clearly seen in a diffuse form in the silver staining test, and no clear bands can be seen, which proves that the protein is in the click reaction process. Destroyed; when adding THPTA (conditions 2, 3, 4), clea...
Embodiment 3
[0148] Embodiment 3: optimization of lysate
[0149] Under different conditions of the lysate (Table 4), other components are the same as in Example 1, and the relative concentrations of the obtained proteins are compared (the comparison between the proteins is not the absolute concentration of itself, that is, no standard is used as the Standard curve for absolute quantification), other steps refer to Example 1. The test results show that the lysate of the present application can enrich RBP more efficiently.
[0150] Table 4 lysate conditions
[0151] Lysate Components Relative protein concentration (mg / ml) 150mM NaCl, 0.1% SDS, 0.5% NP40 0.873 150mM LiCl, 0.1% LiDS 0.738 500mM LiCl, 0.1% LiDS 0.789 500mM LiCl, 0.5% LiDS 0.904 500Mm LiCl, 1% LiDS 0.955
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com