Gene related to bm2 phenotyping, variant and molecular marker thereof
A technology of molecular markers and genes, applied in the field of plant genetic engineering, can solve the problems of loss of MTHFR function and unclear mutation mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Corn bm2 Mutant phenotype and molecular identification
[0026] corn bm2 The mutant seeds were obtained from the mutant library in maizegdb (www.maizegdb.org), and its Stock ID is 114A bm2-PI586725. Use ordinary nutrient soil, seed, cultivate, and continue to observe the phenotype. When the seedlings grow to about the six-leaf stage, the leaf veins appear reddish-brown phenotype ( figure 1 ). Take fresh red-brown leaf vein tissue, and take non-red (B73) leaf vein tissue as a control, use TriZol Reagent (Invitrogen, Cat. No. 15596026) to extract leaf vein total RNA, and use agarose gel electrophoresis and nucleic acid analyzer (NanoDrop) to detect total RNA 2.0 μg of total RNA was used for reverse transcription reaction. The reverse transcriptase used was M-MLV (Promega, Cat. No. M1701). For the steps of reverse transcription reaction, refer to the instructions for use of the reverse transcriptase. The first-strand cDNA synthesized by reverse transcriptio...
Embodiment 2
[0034] Embodiment 2: bm2 mutant detection probe design
[0035] Subsequently, we extracted the genomic DNA (CTAB method) of the control plant (B73) and the mutant plant (114A), and performed conventional PCR amplification using primers ZmMTHFR-proF / R ZMTHFR For the promoter region of the gene, the PCR reaction system is: 2 μL cDNA, 5 μL 10×Buffer, 4 μL dNTP (2.5 mM), forward / reverse primer (10 μM) 1 μL each, 0.5 μL Taq enzyme (5 U / μL ) and 36.5 μL ddH 2 O. Mix well after adding the sample on ice. The PCR reaction conditions are: 94 o C 5 min; 94 o C 30sec, 56 o C 45 sec; 72 o C 2 min, 34 cycles; 72 o C for 10 min. A fragment of about 2.7 kb was obtained by PCR amplification. The primer sequences are as follows:
[0036] ZmMTHFR-proF: TTAGAGTGTGGATAAAATGTACTACC
[0037] ZmMTHFR-proR: CTCGAAAACCTCACCACGAGCTT
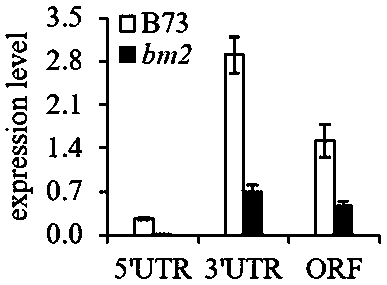
[0038] The results of electrophoresis showed that the amplified fragment in the bm2 mutant was about 400 bp longer than the amplified fragment in B73. Subseq...
Embodiment 3
[0042] Example 3: Effect of MITE transposon on MTHFR transcription level
[0043] Based on the example shown in Example 1 bm2 mutant ZMTHFR Expression level declines and embodiment 2 shows ZMTHFR MITE transposons inserted into the gene promoter region, we analyzed the MITE transposon insertion pairs ZMTHFR Transcription effects. The method used is the detection of promoter transcription activity, and the vector used is pGREEN II 0800-LUC [Hellens et al, Transient expression vectors for functional genomic, quantification of promoter activity and RNA silencing in plants, Plant Methods, 2005, 1:13]. First of all, we will respectively obtain in the embodiment 2 in B73 respectively MTHFR Gene promoter (about 2.7 kb) and bm2 middle MTHFR The gene promoter (about 3.1 kb) was recovered using a gel extraction kit (Promega), and then ligated into the pGREEN II 0800-LUC vector using In-fusion enzyme (abm company) to form B73-5'NCR and bm2 -5' NCR recombinant plasmid, transfor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com