Method for amplifying DNA by using thermophilic primase
A technology of thermal initiation and DNA molecules, which is applied in the field of DNA amplification, can solve the problems of complex operation, multiple proteins, and low sequence specificity, and achieve the effect of good application prospects, high amplification efficiency, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
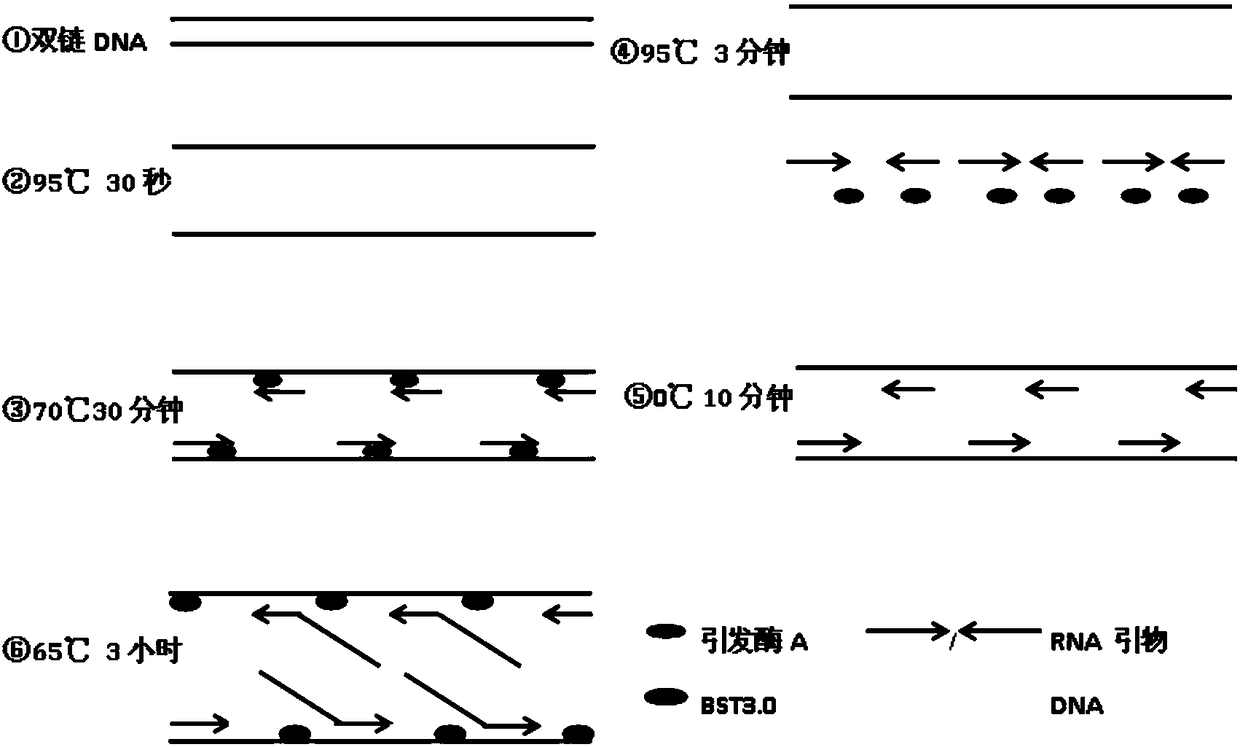
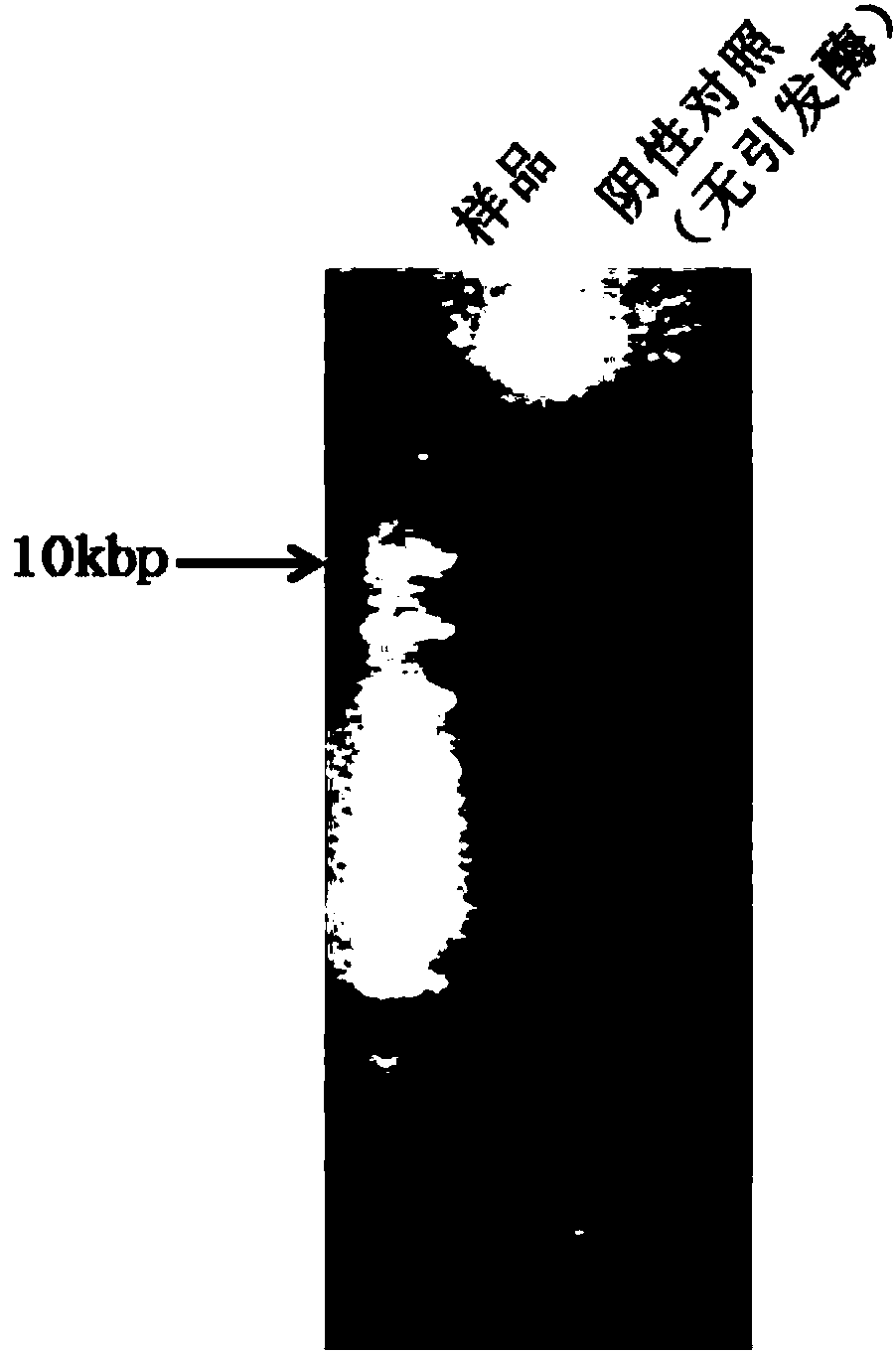
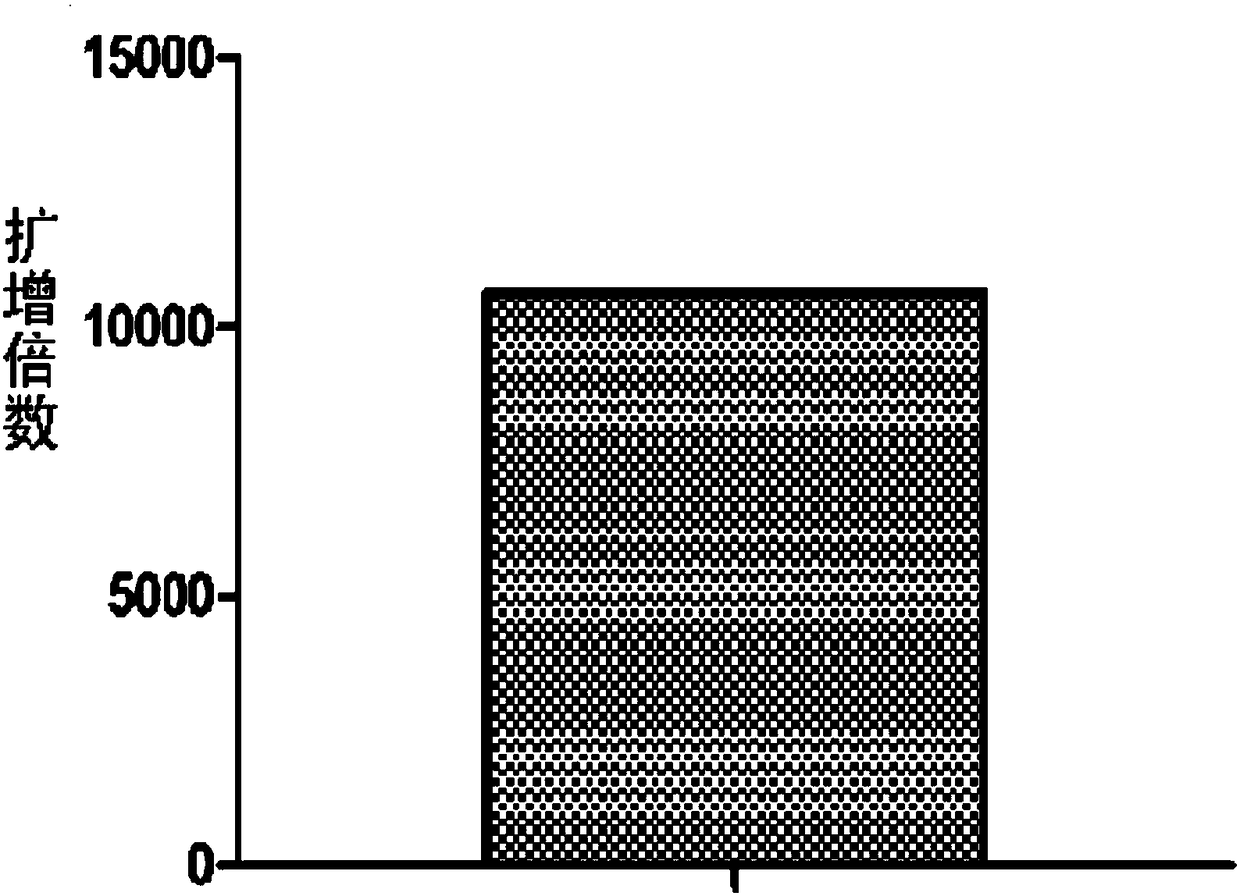
[0038] 5 ng of plasmid pET28a, 50 mM HEPES (pH 7.5), 10 mM DTT, 100 mM potassium glutamate, 5 mM magnesium acetate, 100 μM NTP, 500 μM dNTP, 1 μM thermophilic elicitase A. Prepare 50 μL and mix well, heat at 95°C for 30 seconds, keep at 70°C for 30 seconds, heat at 95°C for 3 minutes, place on ice for 10 minutes, add polymerase BST3.016u, and hold at 65°C for 3 hours. Amplified products were detected by 1% agarose gel electrophoresis. figure 2 , the product molecule is a larger DNA molecule. The spectrophotometer detects the total amount of the DNA product, and calculates the amplification factor. The results show that the method of this embodiment can amplify the nanogram-level template more than 10,000 times, see image 3 .
Embodiment 2
[0040] 5ng circular DNA of a virus genome, 50mM HEPES (pH7.5), 10mM DTT, 100mM potassium glutamate, 5mM magnesium acetate, 100μM NTP, 500μM dNTP, 1μM thermophilic elicitase A. Prepare 50 μL and mix well, heat at 95°C for 30 seconds, keep at 70°C for 30 seconds, heat at 95°C for 3 minutes, place on ice for 10 minutes, add polymerase BST3.016u, and hold at 65°C for 3 hours. Amplified products were detected by 1% agarose gel electrophoresis. Figure 4 , the amplified product is a larger DNA molecule.
Embodiment 3
[0042] 2.5 pg pET28a plasmid DNA, 50 mM HEPES (pH 7.5), 10 mM DTT, 100 mM potassium glutamate, 5 mM magnesium acetate, 100 μM NTP, 500 μM dNTP, 1 μM thermophilic elicitase A. Make 20 μL and mix well, heat at 95°C for 30 seconds, keep at 70°C for 30 seconds, heat at 95°C for 3 minutes, place on ice for 10 minutes, add polymerase 16u, and hold at 65°C for 3 hours. Expand the system to 50 μL, heat at 95°C for 30 seconds, keep at 70°C for 30 seconds, heat at 95°C for 3 minutes, place on ice for 10 minutes, add polymerase BST3.016u, 65°C for 3 hours, and use 1% agar to amplify the product Glucose gel electrophoresis detection, see electrophoresis Figure 5 , resulting in larger bands with irregular smears. The spectrophotometer detects and calculates the content of the product, and the amplification factor exceeds 50 million, see Figure 6 .
[0043] According to the sequence of pET28a, three pairs of detection primers were designed respectively:
[0044] Primer 1 (forward): GG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com