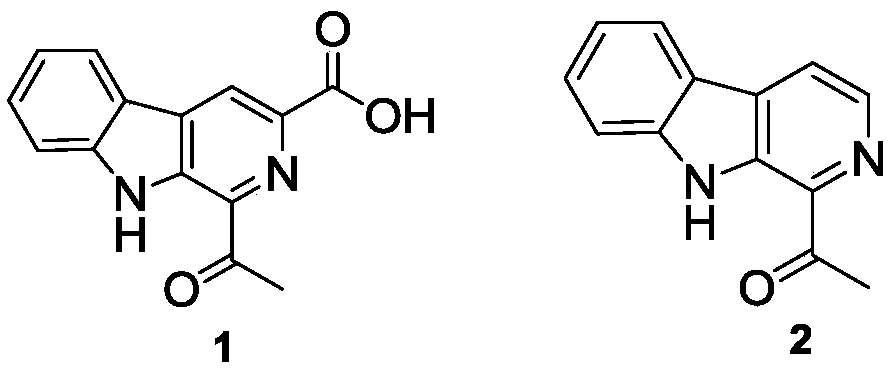
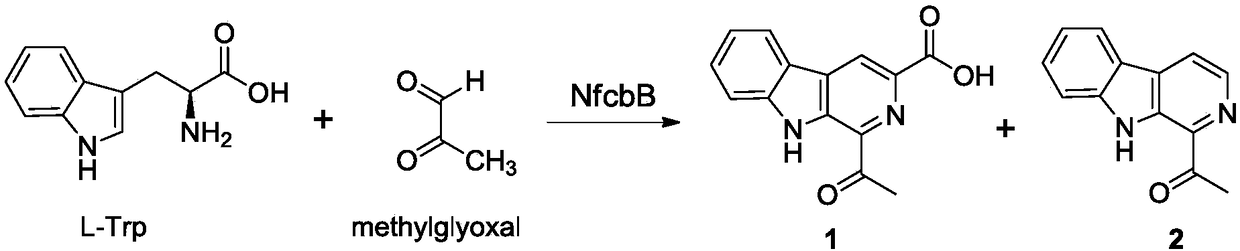
Pictet-Spengler enzyme and encoding gene and application thereof
A technology of encoding gene and transgenic cell line, applied in Pictet-Spengler enzyme and its encoding gene and application field, can solve problems such as restricting application, restricting family enzyme system research, etc., and achieving the effect of enhancing cognition
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Extraction of Actinomycete Nocardiopsis flavescens CGMCC 4.5723 Genomic DNA:
[0049] The fresh spores of actinomycete Nocardiopsis flavescens CGMCC 4.5723 were inoculated into 50 mL of TSB medium (tryptone 17g, plant peptone 3g, sodium chloride 5g, dipotassium hydrogen phosphate 2.5g, glucose 2.5g according to the inoculum size of 5% by volume fraction) , add water to 1L, pH 7.2-7.4), 28-30°C, culture with shaking for about 2 days, and centrifuge at 4000r / min for 10min to collect mycelia. The mycelium was washed twice with STE solution (NaCl 75mmol / L, EDTA 25mmol / L, Tris-HCl 20mmol / L, and the balance was water), and 30mL of STE solution and a final concentration of 3mg / L were added to the washed mycelium. 3 mL of lysozyme, vortex evenly, incubate at 37°C for 3h, add proteinase K to a final concentration of 0.1-0.2mg / mL, mix well, incubate at 37°C for 10min, add SDS to a final concentration of 1-2%, mix well, Put it in a water bath at 55°C for about 1 hour, and invert ...
Embodiment 2
[0051] Expression of NfcbB encoding gene nfcbB in E.coli BL21(DE3):
[0052] Using the genomic DNA of Nocardiopsis flavescens CGMCC 4.5723 as a template, primers were designed. The F sequence of the upstream primer is: CATATGCGCCAGCTAGAGATCGAA, and the restriction site is Nde I; the R sequence of the downstream primer is: AAGCTTTCACCCCCGGGGGTAGGGGTA, and the restriction site is Hind III. The gene nfcbB was amplified by PCR, and the PCR reaction system was: (50 μL system): 5×Fast Pfu Buffer 10 μL, dNTP Mix 5 μL, upstream primer F (10 μM) 1.5 μL, downstream primer R (10 μM) 1.5 μL, DMSO 3 μL, Template 10ng, Fast Pfu DNA Polymerase 0.5μL, ddH 2 O 27.5 μL. The reaction conditions and cycle parameters were: 95°C pre-denaturation for 5 min; 95°C denaturation for 50 sec, 60°C for 50 sec, 72°C for 1.5 min, 30 cycles; 72°C extension for 10 min. The obtained PCR product was sequentially recovered by gel, adding an "A" tail, TA cloning, and sequencing verification (the nucleotide seque...
Embodiment 3
[0054] Detection of fermentation and metabolites of recombinant strain E.coli BL21(DE3) / pET28a(+) / nfcbB:
[0055] Streak culture the obtained recombinant expression strain E.coli BL21(DE3) / pET28a(+) / nfcbB onto LB solid plate, pick a single clone after overnight culture at 37°C, and inoculate at an inoculum size of 1% by volume fraction Put 50mL of LB culture liquid into a 250mL Erlenmeyer flask, and culture it on a shaker at 28°C at 180r / min until OD 600 At about 0.6, add isopropyl-β-D-thiogalactopyranoside (IPTG) at a final concentration of 0.1 mM to the culture medium. After continuing to incubate at 28°C for 12 hours, add an appropriate amount of 1 mol / L HCl to the fermented product to adjust its pH to 5, add twice the volume of ethyl acetate, and sonicate for 5 minutes to disrupt the cells, and then let stand to separate. The ethyl acetate extract was separated from the water phase, and the ethyl acetate was evaporated to dryness with a rotary evaporator, and the residue ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com