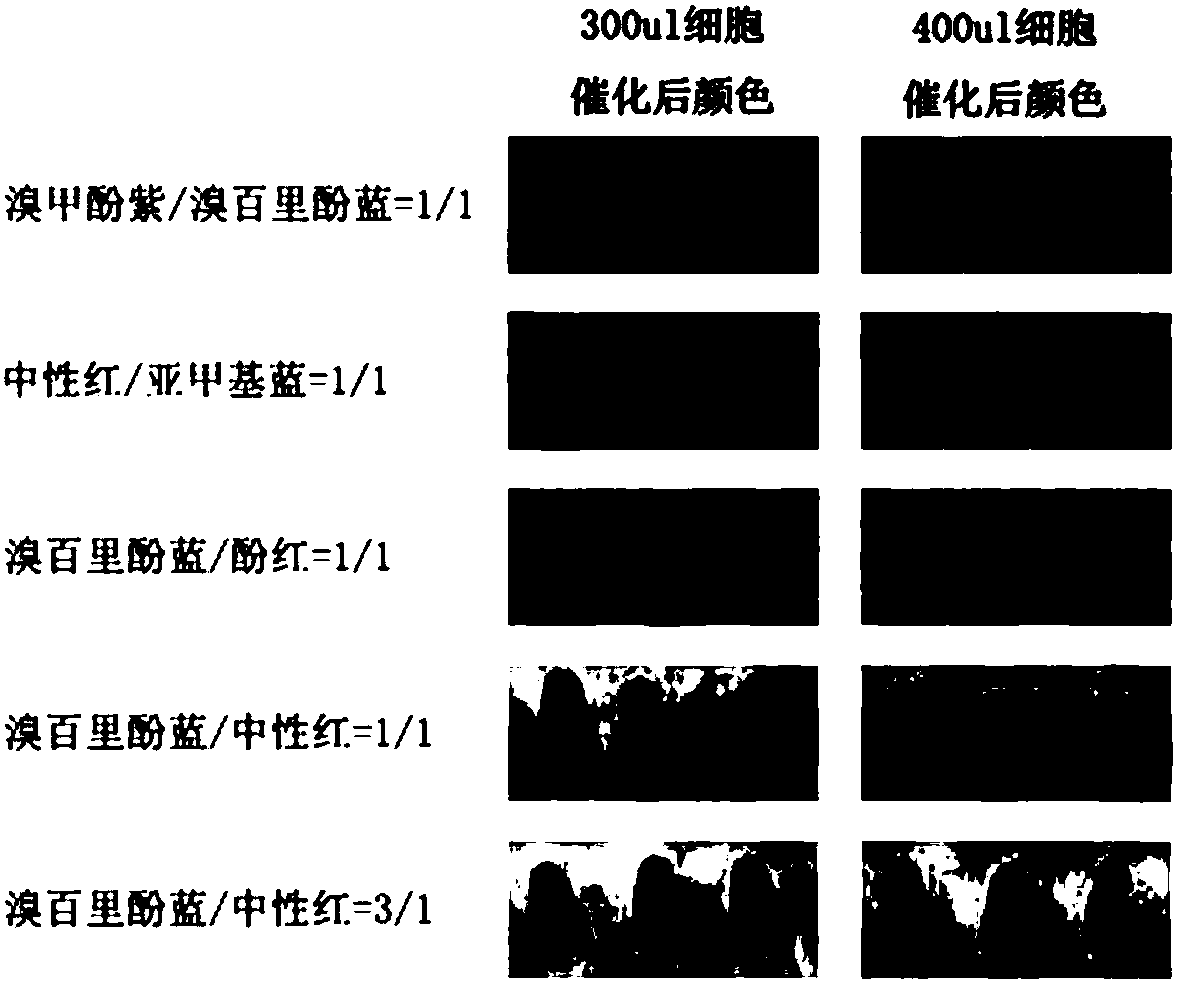
Lysine decarboxylase mutant library construction method and high-throughput screening method of high-activity or low-activity mutant strains
A technology of lysine decarboxylase and construction method, which can be used in microorganism-based methods, chemical libraries, biochemical equipment and methods, etc., and can solve problems such as difficult amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1. Construction of Escherichia coli Lysine Decarboxylase CadA Expression Vector
[0057] (1) Amplification of CadA gene
[0058] Escherichia coli KG1665 (Beijing Tianenze Biotechnology Co., Ltd.) was cultured overnight in LB medium (37°C, 220rpm), and 4ml of bacterial solution was taken the next day, and the genome extraction kit (Axygen, AP-MN-P-250G) ) Extract its genome. The primers CadAF 5'-GGAATTCCATATGATGAACGTTATTGCA ATAT-3' (SEQ ID NO:1) and CadAR 5'-CCCAAGCTTCCTTGTACGAGCT AATTAT-3' (SEQ ID NO: 2) designed according to the gene sequence of E. coli lysine decarboxylase CadA were used to Genome (GenBank: U00096.3) was used as a template to amplify the CadA gene fragment. Design NdeI and HindIII restriction sites on the amplification primers. The amplification method is a conventional PCR method, and the DNA polymerase used is from Takara Bioengineering (Dalian) Co., Ltd. (TAKARA) Max DNA polymerase.
[0059] The amplification system is as follows:
[0060] ...
Embodiment 2
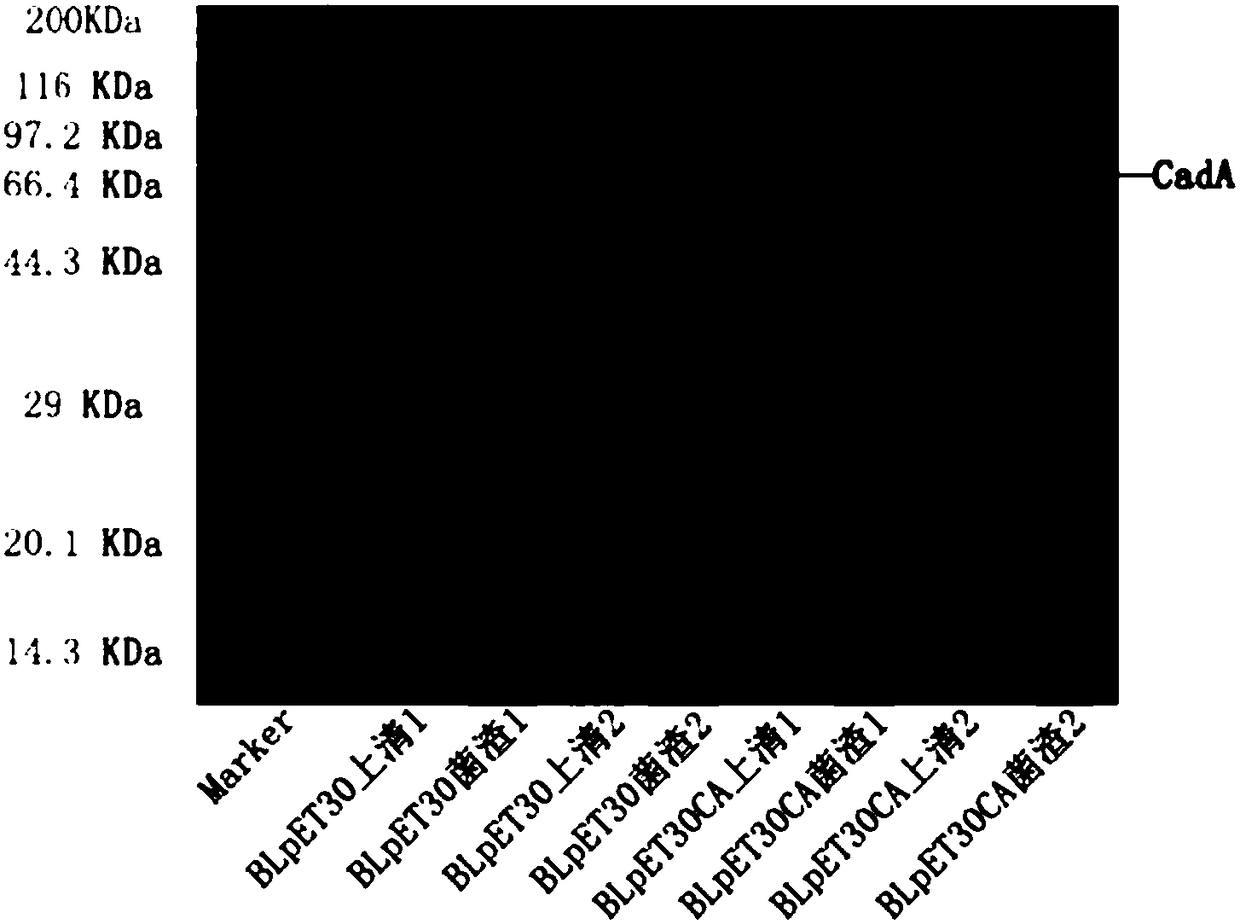
[0071] Example 2. Induced expression of CadA
[0072] (1) Transform the plasmid pET30CA into E. coli BL21 (DE3) to obtain the strain BLpET30CA. The empty plasmid pET30 was transformed into BL21(DE3) to obtain the control strain BLpET30.
[0073] (2) Inoculate control BLpET30 and BLpET30CA into 5ml test tubes of LB liquid medium (containing kanamycin 50mg / ml), inoculate two tubes each, and cultivate overnight in a shaker at 37°C and 220rpm.
[0074] (3) Transfer the bacterial solution from step (2) overnight culture to a new 5ml liquid LB medium (containing kanamycin 50mg / ml) at a transfer volume of 2% (v / v), and continue at 37 Cultivate in a shaker at 220 rpm.
[0075] (4) Cultivate to OD 600 When it reached 0.6, IPTG was added to induce expression, and the final concentration of IPTG was 0.1 mM. Continue to incubate in a shaker at 37°C and 220 rpm.
[0076] (5) Collect 1ml of bacterial solution and centrifuge after 6 hours of inducing expression. Collect the supernatant and bacteri...
Embodiment 3
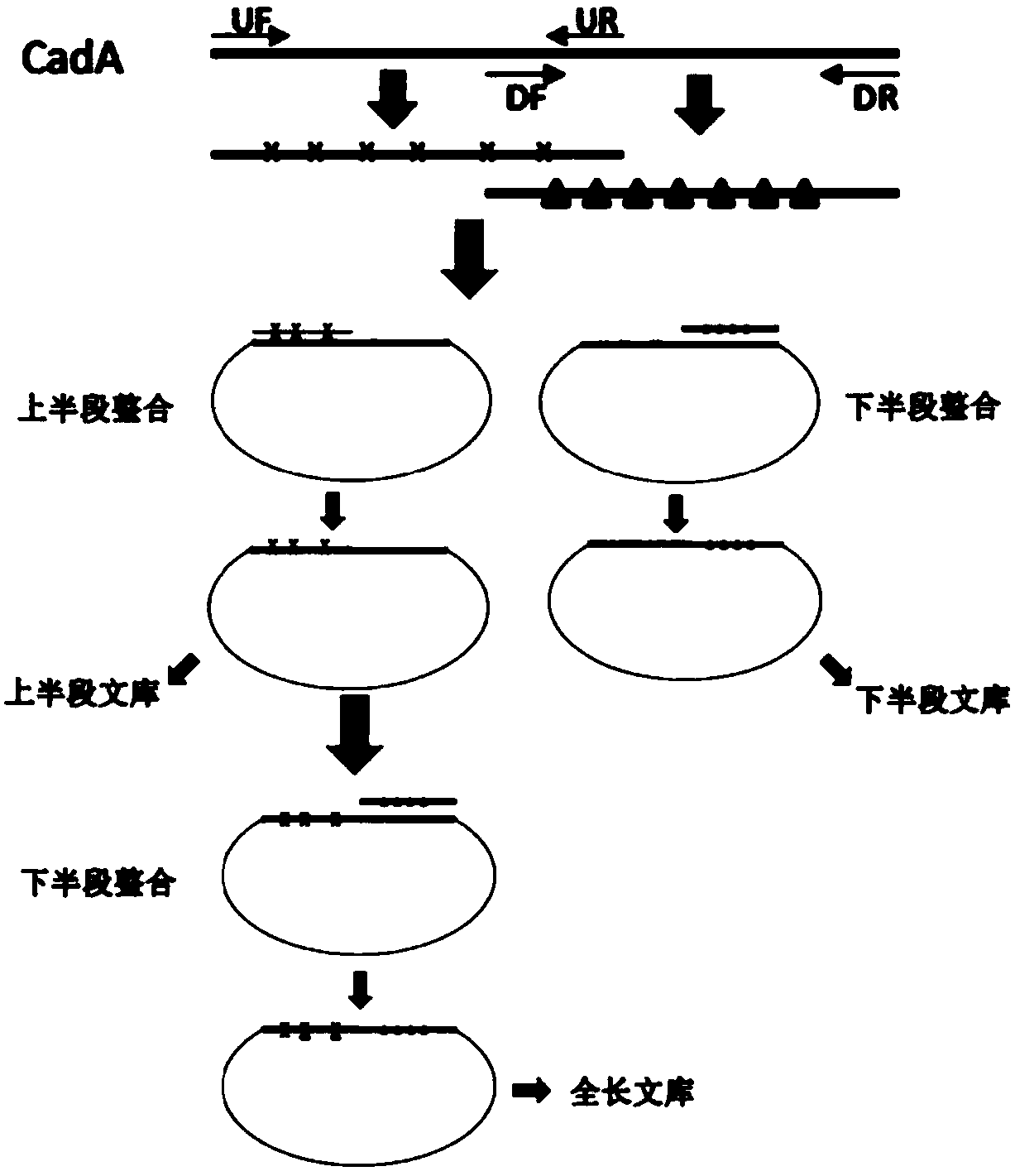
[0080] Example 3. Construction of mutant library
[0081] To build a library of CadA segmented mutations, according to the length, the 800-1200bp after the start codon of CadA ATG is divided into the "upper half", and the 800-1200bp before the stop codon TAA of CadA is divided into the "lower half" Segment"; According to function, the plp binding region (552-1241bp) in CadA is divided into "plp segment". Design mutant primers UF 5'-TGTTTAACTTTAAGAAGGAGA-3' (SEQ ID NO: 3), UR 5'-TCTTCGTTTACGTCACCT-3' (SEQ ID NO: 4) according to the gene sequence information of the upper, lower and plp segments; DF 5'- CCAACTTCTCACCGATTTA-3' (SEQ ID NO: 5), DR 5'- TCAAGACCCGTTTAGA GGC-3' (SEQ ID NO: 6); plpF 5'-GTAGCCTGTTCTATGATTTCTT-3' (SEQ ID NO: 7), plpR 5'-TTACCT GCATTGCCTTTC-3' (SEQ ID NO: 8). Add the following reagents to the PCR tube: 5ul of 10×rTaq Buffer (Takara), 8ul of dNTPx (2.5mM dATP, 1.25mM dTTP, 1.25mM dCTP, 2.5mM dGTP), 1.5ul UF or DF or plpF, 1.5ul UR or DR or plpR (UR with UF,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com