Tripterygium wilfordii triterpene synthase twosc3 and its coding gene and application
A coding and amino acid technology, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of restricted development, slow growth of plants, and low content.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Embodiment 1, the cloning of Tripterygium wilfordii Twosc3 full-length cDNA sequence
[0057] 1. Extraction of total RNA from tripterygium wilfordii suspension cells and first-strand cDNA acquisition
[0058] Using improved CTAB method (CTAB Buffer: 2% CTAB (W / V); 100mmol L -1 Tris-HCl (pH 8.0); 25mmol L -1 EDTA; 2.0mol L -1 NaCl; 0.5g L -1 spermidine) to extract the total RNA of Tripterygium wilfordii suspension cells. The obtained total RNA was used as a template, and the 5'-CDSprimer in the SMARTerTM RACE cDNA Amplification Kit was used to perform reverse transcription to obtain 5'-RACE-Ready cDNA.
[0059] 2. Primer Design
[0060] The full-length sequence fragment of the gene was screened according to the transcriptome data annotation of Tripterygium wilfordii, and Twosc3-F and Twosc3-R primers were designed. The primer sequences are as follows:
[0061] Twosc3-F: AAACACAAGAGATTGATATAAGC (SEQ ID NO: 3)
[0062] Twosc3-R: TTCGATTTAGCGGACACT (SEQ ID NO: 4)
...
Embodiment 2
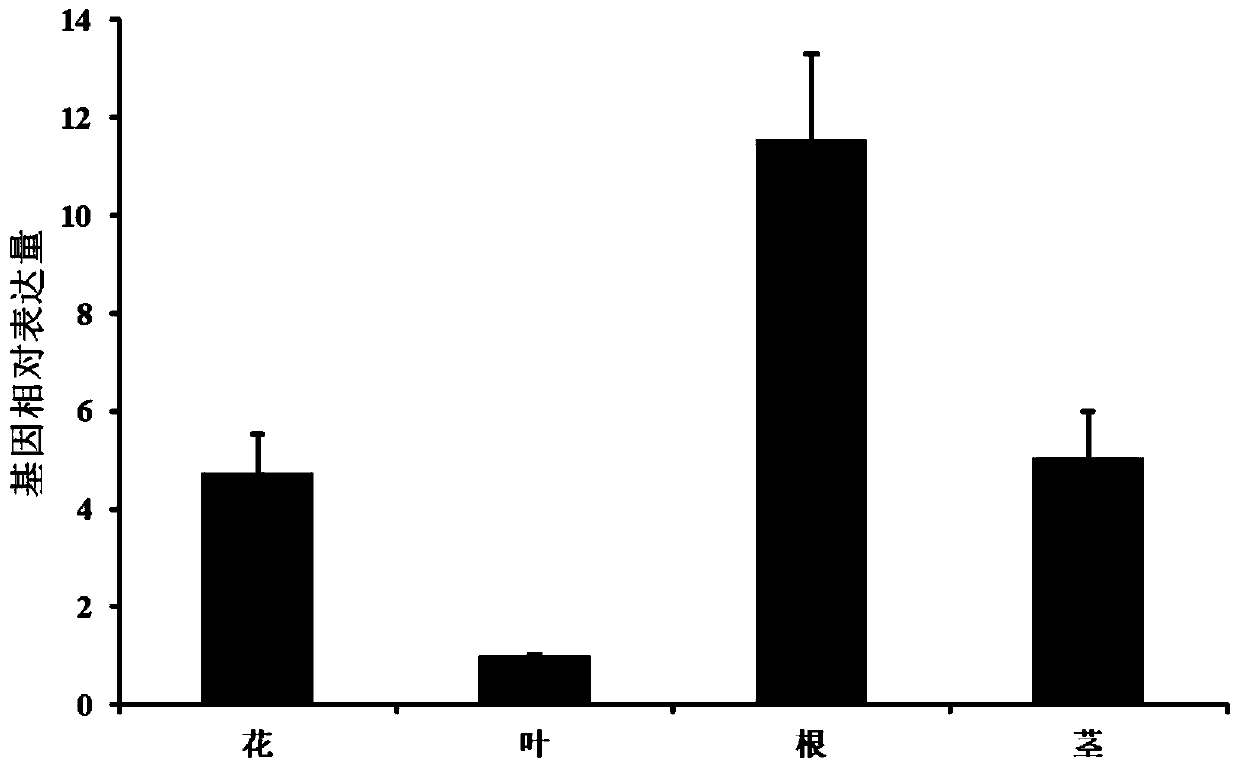
[0092]Embodiment 2, Twosc3 gene tissue expression analysis
[0093] 1. Handling of Experimental Materials
[0094] The roots, stems, leaves, and flowers of Tripterygium twig are collected from five different plants in the Yongan State-owned Forest Farm in Fujian. The samples were taken back to the laboratory for cleaning, quick-frozen in liquid nitrogen and stored in a minus 80 refrigerator.
[0095] 2. Extraction of total RNA and real-time quantitative PCR
[0096] The roots, stems, leaves, and flowers stored in the negative 80 refrigerator were crushed in a liquid nitrogen environment, and the total RNA was extracted by the improved CTAB method, and reverse-transcribed into cDNA with the FastQuant RT Kit (Tiangen). as an internal reference. Real-time quantitative PCR was performed using ABIPrism 7300 Sequence Detection System (Applied Biosystems, USA) and KAPA SYBR FASTUniversal 2X qPCR Master Mix (Kapa Biosystems, USA) kit. Real-time fluorescence quantitative reaction s...
Embodiment 3
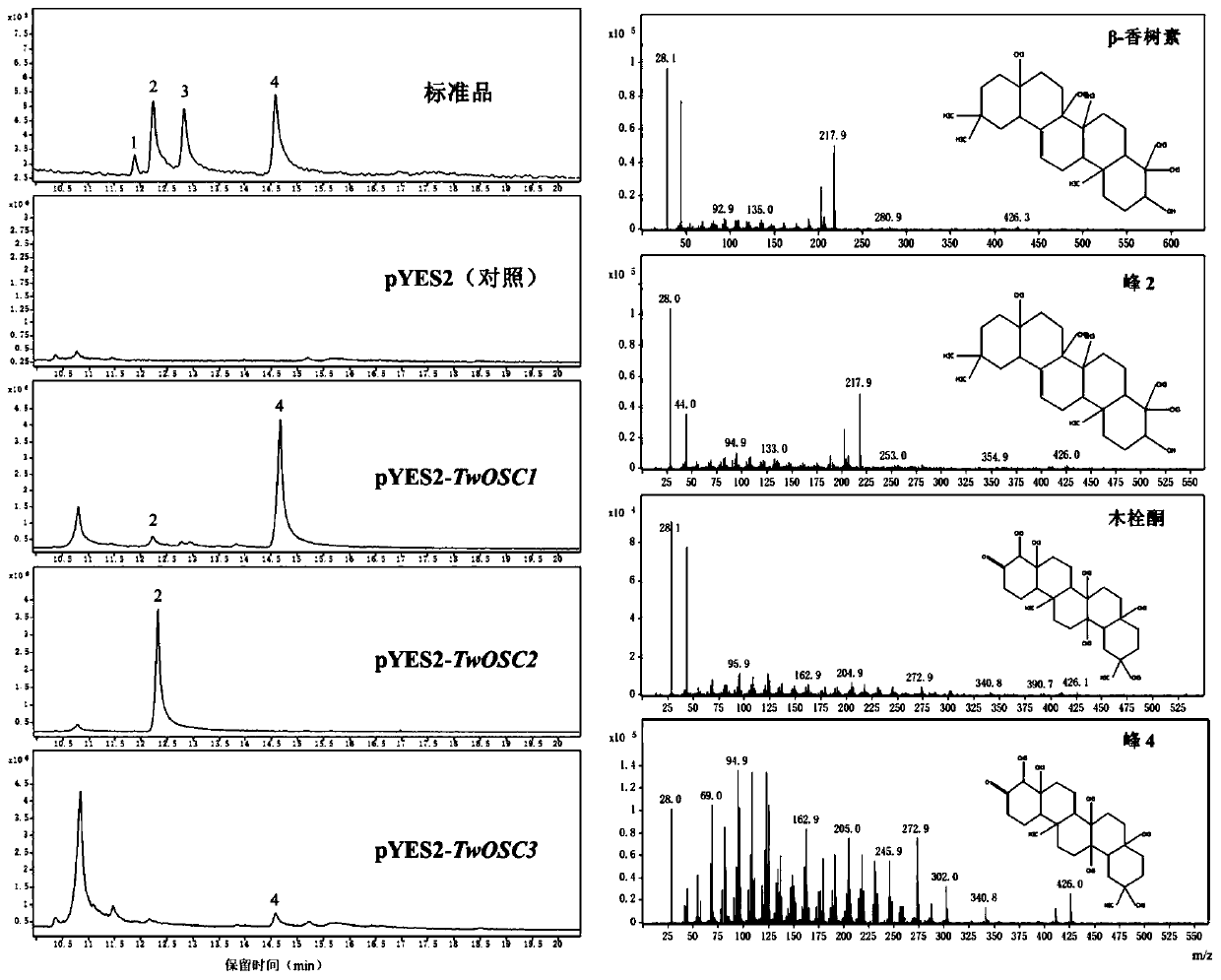
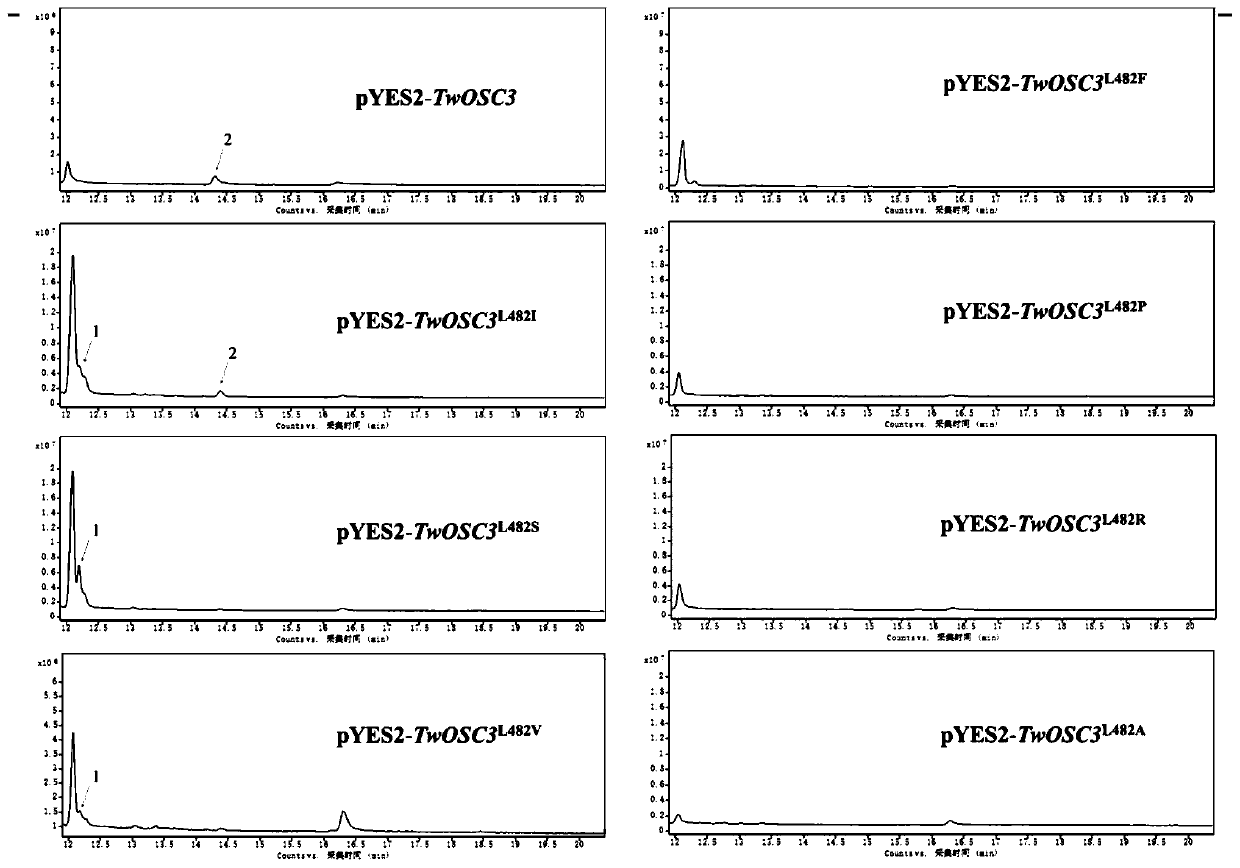
[0102] Example 3, research on the biological function of Tripterygium wilfordii Twosc3
[0103] 1. Construction of eukaryotic expression vector
[0104] The vector pEASY-Blunt-Twosc3 plasmid containing the full-length cDNA of Tripterygium wilfordii Twosc3 gene was used as a template, and the gene coding region was amplified by PCR with primers containing restriction sites (marked with a horizontal line is the restriction site). DNA polymerase adopts high-fidelity DNA polymerase (Phusion High-Fidelity PCR Master Mix). The PCR parameters were 98°C for 30s, 1 cycle; 98°C for 10s, 60°C for 10s, 72°C for 2min 30s, 35 cycles; 72°C for 5min; 4°C for maintenance. The amplified product was recovered by Gene JET GelExtraction Kit gel (method as follows).
[0105] Twosc3-F:CGG GGTACC ATGTGGAAGATAAAGATAGCTGA (SEQ ID NO: 9)
[0106] Twosc3-R:CGC GGATCC CTATTTAATTTCCTTTGAAGGC (SEQ ID NO: 10)
[0107] Gene JET Gel Extraction Kit glue recovery step (with the glue recovery step in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com