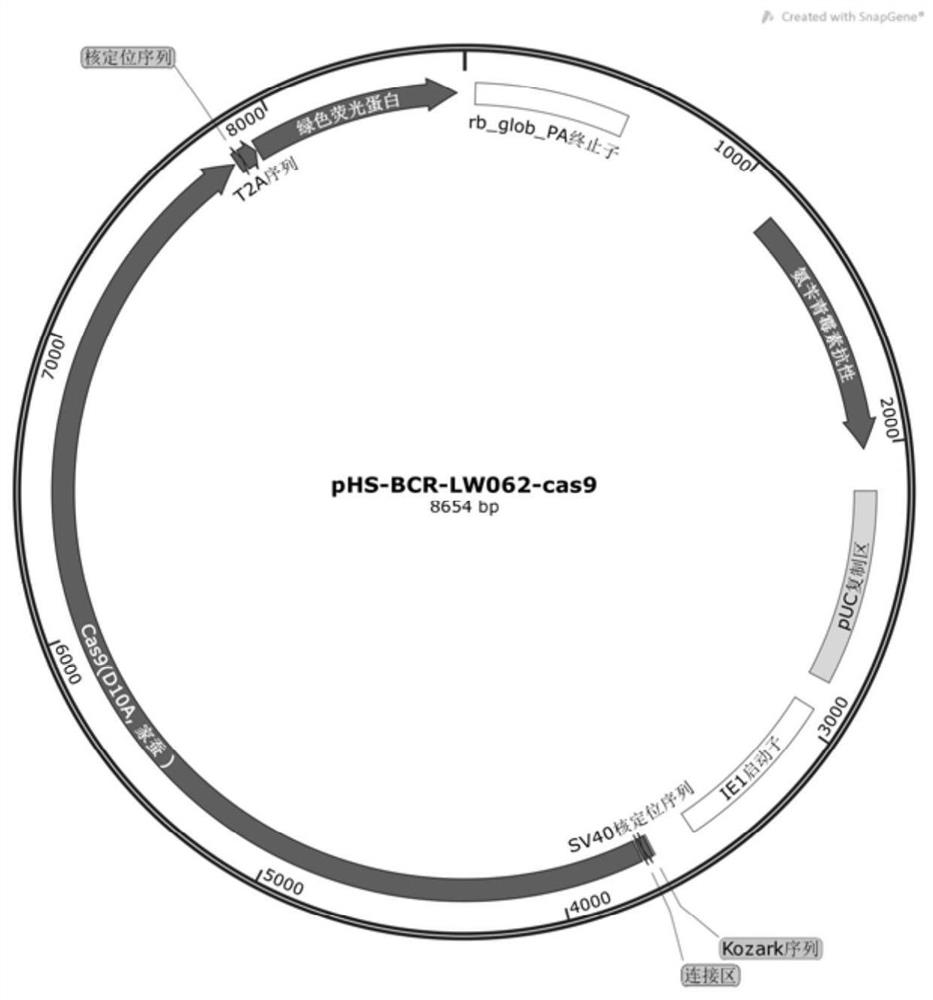
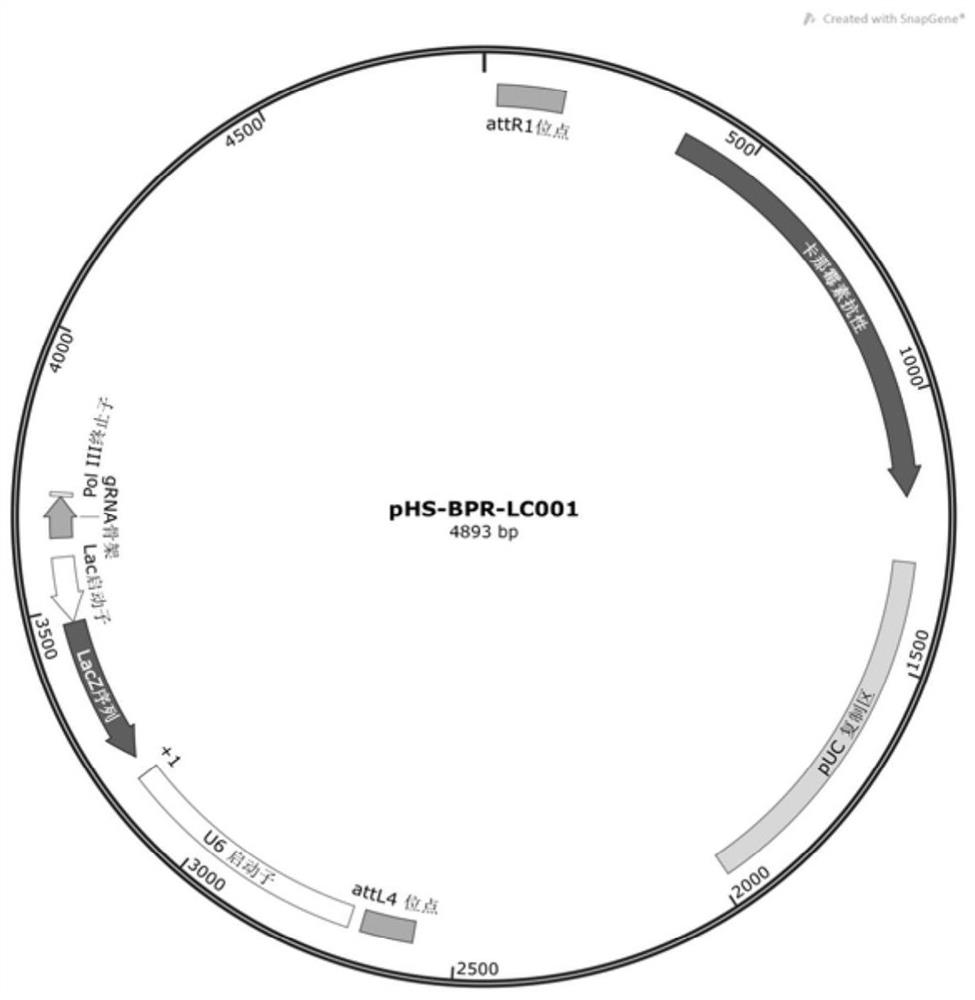
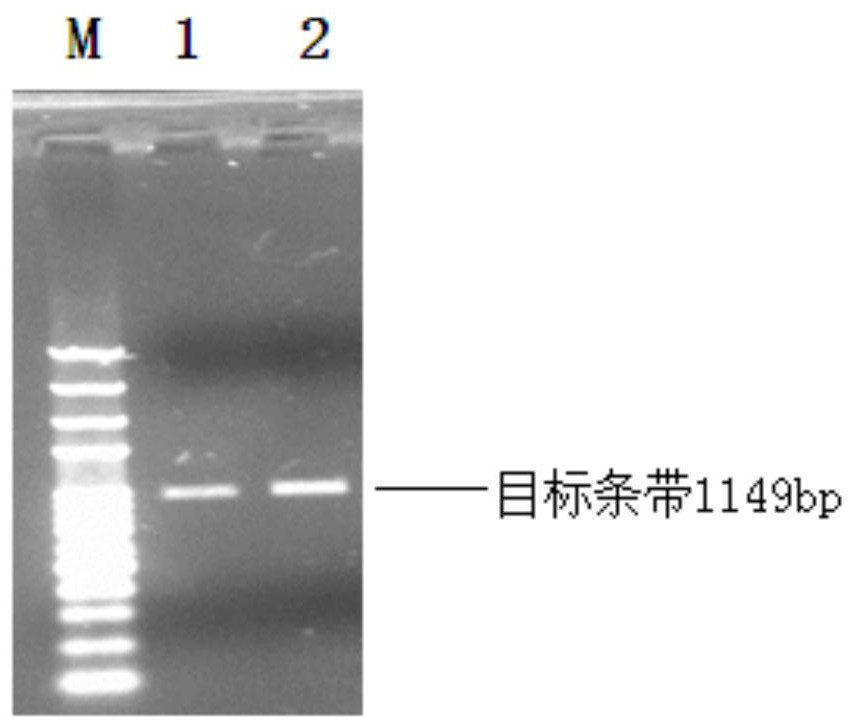
Silkworm gene precise knockout system based on CRISPR/cas9 double nickase technology and its application
A nicking enzyme, phs-bcr-lw062-cas9 technology, applied in the field of molecular biology, can solve the problems of low specificity, CRSIPR/Cas9 off-target mutation, etc., and achieve the effect of simple reaction process, reduced off-target rate, and increased expression efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Vector construction and preparation of knockout system
[0025] Taking the silkworm BmBLOS2 gene as an example, the test steps are as follows:
[0026] 1 Design and selection of targeting gRNA
[0027] The BmBLOS2 gene sequence was found in the silkworm genome database, and the upstream and downstream regions of the first and second exons of the gene were selected to design target sites. The gRNA sequence was designed by the online design tool (http: / / crispr.dbcls.jp), and sticky ends were added to both ends of the gRNA sequence. The primer sequences were designed as follows:
[0028]
[0029] Reverse primer → NNNNNNNNNNNNNNNNNNNNCAAA
[0030] A site needs a pair of gRNAs, and the PAMs of each other can be combined head-to-tail, head-to-head or tail-to-tail, with an interval of about 4-20bp. The selected target site sequences are shown in Table 1:
[0031] Table 1 Targeting gRNA sequences
[0032]
[0033] 2 gRNA synthesis and vector construction
...
Embodiment 2
[0058] Example 2 Targeted knockout of the silkworm BmBLOS2 gene
[0059] The knockout system established in Example 1 was used to construct site-directed knockout vectors of the corresponding genes, respectively, and transfected silkworm ovary cells (BmN).
[0060] 1 BmN cell line pre-experiment
[0061] 1.1 Mycoplasma detection: Aspirate 1 mL of BmN cell culture medium supernatant, use mycoplasma-specific primers for PCR amplification, and use agarose gel electrophoresis to identify whether there is mycoplasma contamination in the cell line;
[0062] 1.2. Determination of growth rate: BmN cells were subcultured to observe the growth state and proliferation rate of BmN cells.
[0063] 2 Transfection
[0064] 2.1 Cell plating: Press 10 for cells before transfection 5 Cells / mL were plated until the cell confluence was 60-80% the next day.
[0065] 2.2 Dilute 0.5 μg of plasmid pHS-BCR-LW062 with 25 μL of Opti-MEM medium and mix thoroughly;
[0066] 2.3 Dilute 1.25 μL of Entr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com