Construction method and application of annular RNA overexpression system for protoplast of secondary xylem of poplar
A protoplast and construction method technology, applied in the field of genetic engineering, can solve the problems of low genetic transformation efficiency and long cycle, and achieve the effect of simple and fast construction method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Extraction of poplar nucleic acid
[0038] (1) Material handling
[0039] Select 6 Populus trichocarpa growing healthily at the age of 3-9 months, use the xylem below the seventh leaf on the upper end of the morphology and above the soil surface at the lower end as materials, scrape the secondary xylem with a scalpel after peeling, and put it in Put them in the tin foil that has been marked in advance, and immediately put them in liquid nitrogen for quick freezing. Use a high-throughput tissue disruptor (QIAGEN TissueLyser II) to grind the xylem material into a powder sample, mix it, put it in an RNase-free 50 mL tube (corning), and store it in a -80°C refrigerator for a long time.
[0040] (2) Extraction of xylem Genomic DNA and Total RNA
[0041] Aliquot 120 mg of the powder samples obtained from the treatment of the above materials into 1.5 mL RNase-free tubes, use the PlantGenomic DNA Kit (TIANGEN, no. DP305, China) to extract the Genomic DNA of the xyle...
Embodiment 2
[0044] Example 2 Molecular biological verification of circular RNA
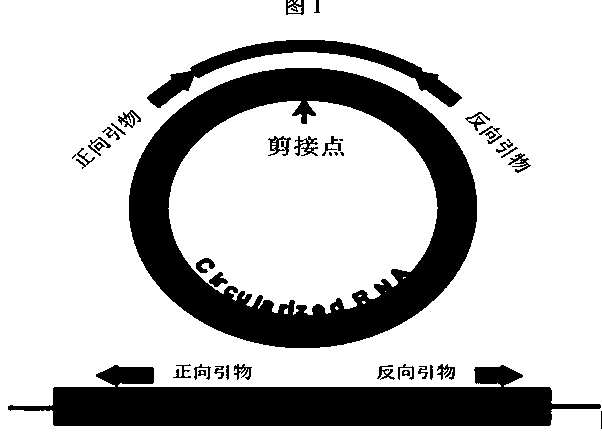
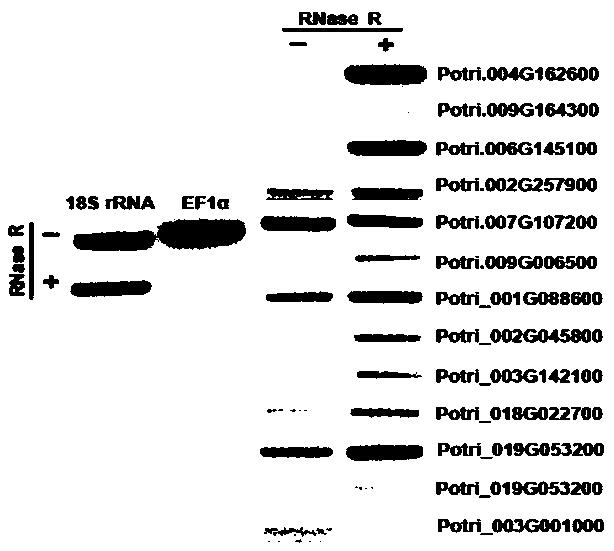
[0045] Through transcriptome sequencing, 13 circular RNA genes were obtained, the sequences of which are shown in SEQ ID NO: 1 to SEQ ID NO: 13. In order to verify the authenticity of the circular RNA, PRAPI software was used to design "back-to-back" primers ( figure 1 ), verified by PCR amplification and sequencing experiments, and the primer sequences are shown in Table 2.
[0046] Take 20 μg Total RNA (54 μL) and divide it equally into two 1.5 mL RNase-free centrifuge tubes, one for RNase R digestion and the other for control. Next, add 3.1 μL 10×RNaseR Reaction Buffer and 1 μL RNase R (20U / uL) to both the treatment tube and the control tube in sequence (in the control tube, 1 μL RNase-free water was used instead of RNase R), mix well, microcentrifuge, and place in a water bath Incubate at 37°C for 10 min. Then, add 30 μL of phenol-chloroform-isoamyl alcohol=25:24:1 to terminate the enzyme digestion rea...
Embodiment 3
[0050] Example 3 Construction of circular RNA overexpression vector
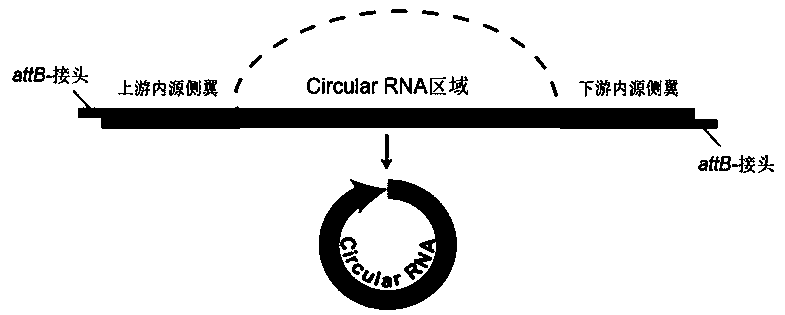
[0051] (1) A band containing the upstream and downstream endogenous flanking sequences of the host gene attB- Frame acquisition of linker sequences
[0052] The partial sequence of the host gene of Potri.004G162600 (Chr04:18369599-18370055) was selected as the target gene. Design 5' end band attB- site linkers, and primers that include the endogenous flanking sequences of the gene of interest ( image 3 ). The primer sequences are as follows:
[0053] The forward primer is, SEQ ID NO:44:
[0054] Ptr-trans-130-F:5'- GGGGACAAGTTTGTACAAAAAAGCAGGCTT
[0055] CGTATATGCAGGCCTGTCTTTGCT-3'
[0056] The reverse primer is, SEQ ID NO:45:
[0057] Ptr-trans-130-R:5'- GGGGACCACTTTGTACAAGAAAGCTGGGTC
[0058] CTAAACAGAAGACGGACACAATTTTGT-3'
[0059] The underlined part is the recognition site of Gateway, namelyattB- recognition site.
[0060] 200 ng Genomic DNA as a template, 2.5 μL each of the above forward...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com