A functional nucleic acid fluorescent sensor and its application in lead ion detection
A fluorescent sensor and functional nucleic acid technology, applied in fluorescence/phosphorescence, instruments, measuring devices, etc., can solve the problems of high detection limit, expensive instruments, high background, etc., achieve high temperature stability, fast and accurate detection, high detection The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] In this embodiment, the preparation of lead ion-dependent deoxyribozyme solution, the steps are as follows:
[0051] Add 3 μL of 2 μM DNAzyme substrate chain stock solution and 6 μL of 0.5 μM DNAzyme enzyme chain stock solution to 2 μL buffer (500 mM HEPES, 500 mM NaCl, 50 mM MgCl 2 ,pH=7.26), then dilute to 18μL with ultrapure water, shake well, heat at 80°C for 3min, then slowly cool down to room temperature, the whole cooling process lasts for 40min, and then stand at room temperature for 25min to obtain lead ion-dependent deoxidation Ribozyme solution. The nucleotide sequence of the DNAzyme substrate chain is shown in SEQ ID No.1 in the sequence listing, and the nucleotide sequence of the DNAzyme enzyme chain is shown in SEQ ID No.2 in the sequence listing.
[0052] Repeat the above operations to prepare several portions of lead ion-dependent DNAzyme solutions, each 18 μL.
Embodiment 2
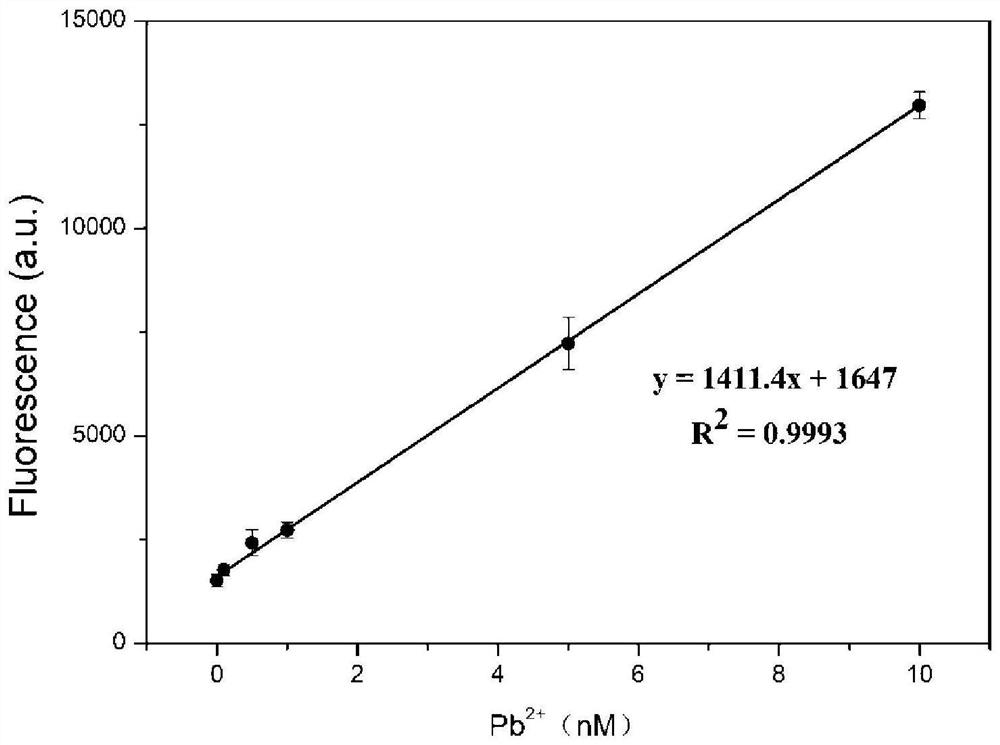
[0054] In the present embodiment, the standard solution curve is drawn, and the steps are as follows:
[0055] (1) Prepare lead ion standard solution
[0056] Preparation of Pb 2+ Lead acetate standard solutions with concentrations of 0, 0.8, 4, 8, 40, and 80 nM, respectively.
[0057] (2) Draw a standard curve
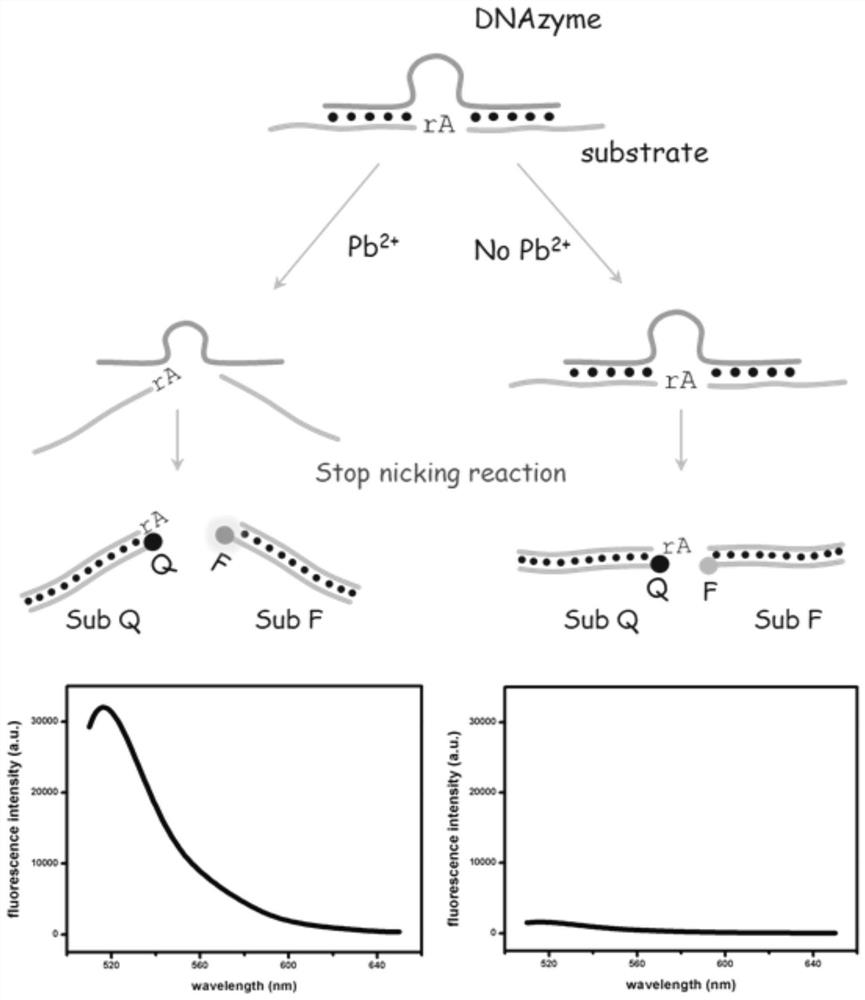
[0058] ① Get a part (18 μL) of the lead ion-dependent deoxyribozyme solution prepared in Example 1, add 2 μL Pb to it 2+ Lead acetate standard solution with a concentration of 0nM, placed at room temperature (25°C) for 80min for cleavage reaction, the cleavage reaction will give two cleavage products with nucleotide sequences as shown in 7 and 8, and then add 2μL of 5' with a concentration of 2μM Sub F labeled with a fluorescent dye group at the end and 4 μL of Sub Q labeled with a fluorescent quencher at the 3’ end at a concentration of 2 μM were mixed and incubated at 37°C for 35 min to terminate the cleavage reaction to obtain a reaction mixture, which was then ...
Embodiment 3
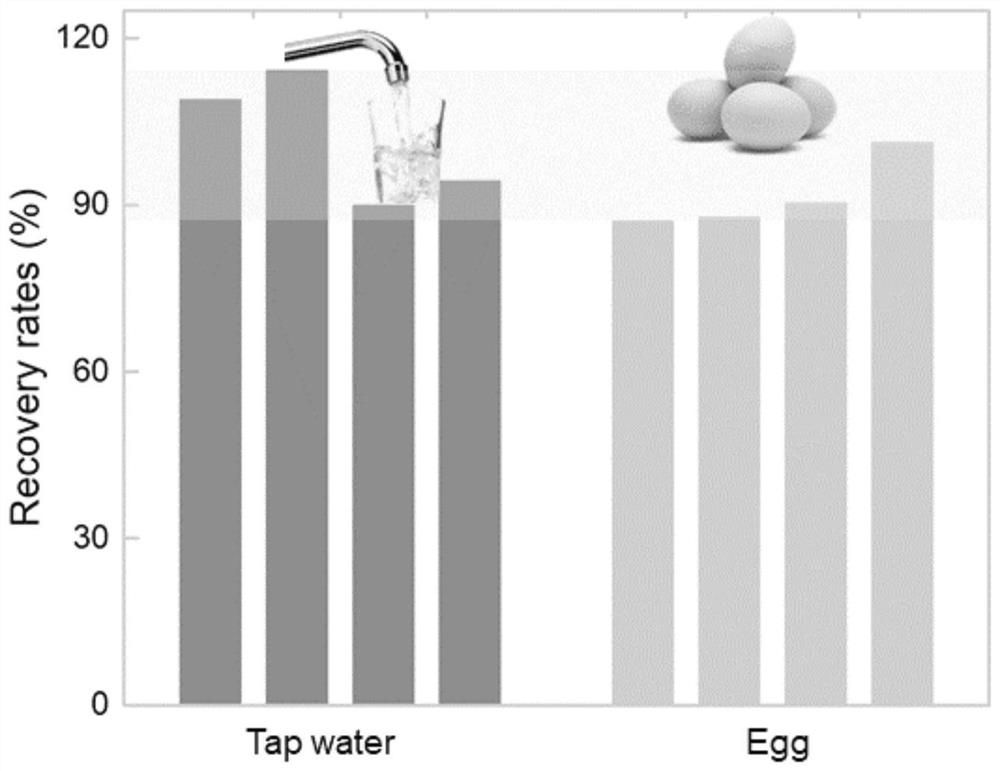
[0061] In this example, adding Pb to tap water 2+ And detect Pb 2+ Concentration, the steps are as follows:
[0062] (1) Prepare Pb from tap water 2+ Concentrations of 0, 4, 8, 40, 80nM Pb 2+ The sample solution is recorded as 1#~5# sample solution in turn.
[0063] (2)Pb 2+ Sample solution detection
[0064] ① Take a portion (18 μL) of the lead ion-dependent deoxyribozyme solution prepared in Example 1, add 1# sample solution to it, and place it at room temperature (25° C.) for 80 minutes to carry out the cleavage reaction. The cleavage reaction will give a nucleotide sequence such as The two cleavage products shown in 7 and 8, and then add 2 μL of 2 μM Sub F labeled with a fluorescent dye at the 5’ end and 4 μL of 2 μM Sub Q labeled with a fluorescent quencher at the 3’ end, After mixing, incubate at 37°C for 35 min to terminate the cleavage reaction to obtain a reaction mixture, and then use a fluorescence spectrometer to measure the fluorescence intensity of the obta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com