Application of physcomitrella patens ATG3 gene in plant senescence and salt tolerance stress
A Physcomitrella patens and gene technology, applied in the application field of Physcomitrella patens ATG3 gene in plant senescence and salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
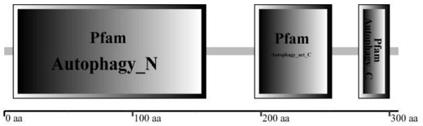
[0019] In the Plant Genome Database (http: / / phytozome.jgi.doe.gov / pz / portal.htmL) BLASTp comparison of the reported Arabidopsis ATG3 protein sequence and download the homologous ATG3 protein in Physcomitrella patens And gene sequence, through the online structure domain analysis tool SMART (http: / / smart.embl.de / ) and InterProScan (http: / / www.ebi.ac.uk / interpro / ) to obtain the candidate gene conservative domain identification, identification results such as figure 1 As shown, the transcribed and translated protein has three structural domains, which are the Autophagy_N domain at the N-terminus, the Autophagy_act_C domain at the middle, and the Autophagy_C domain at the C-terminus. It can be used as a candidate gene, and the number of the Physcomitrella patens ATG3 gene is Pp1s62_116V6.1.
Embodiment 2
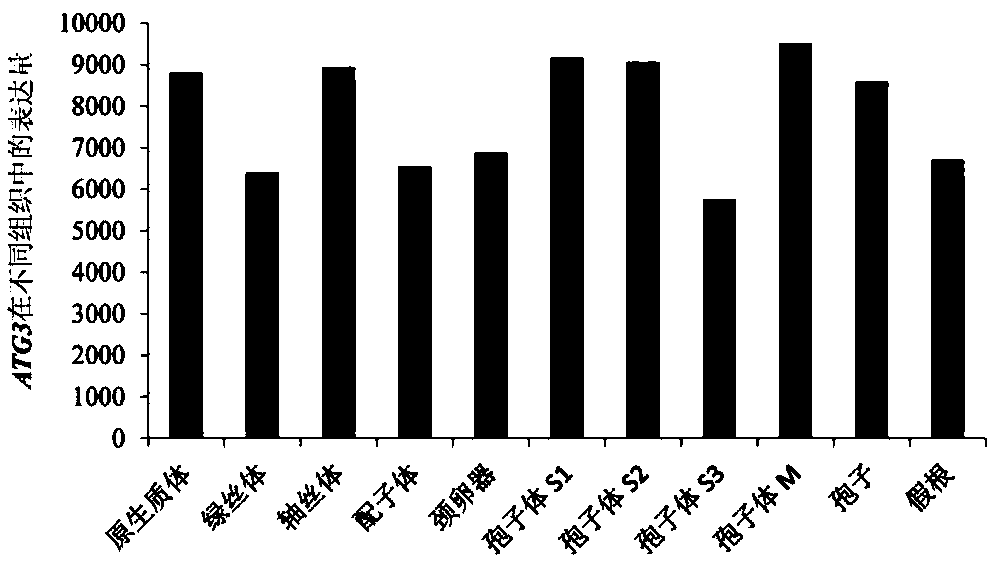
[0021] The Physcomitrella patens ATG3 gene numbering Pp1s62_116V6.1 obtained in Example 1 is input into the online analysis website http: / / bar.utoronto.ca / efp_physcomitrella / cgi-bin / efpWeb.cgi to obtain ATG3 genes different in Physcomitrella patens. The expression levels in tissues, including protoplast, chlorocelium, axoneme, gametophyte, archegonia, sporophyte S1, sporophyte S2, sporophyte S3, sporophyte M, spores and rhizoids. The expression measurement results were as figure 2 As shown, it is expressed in many tissues.
Embodiment 3
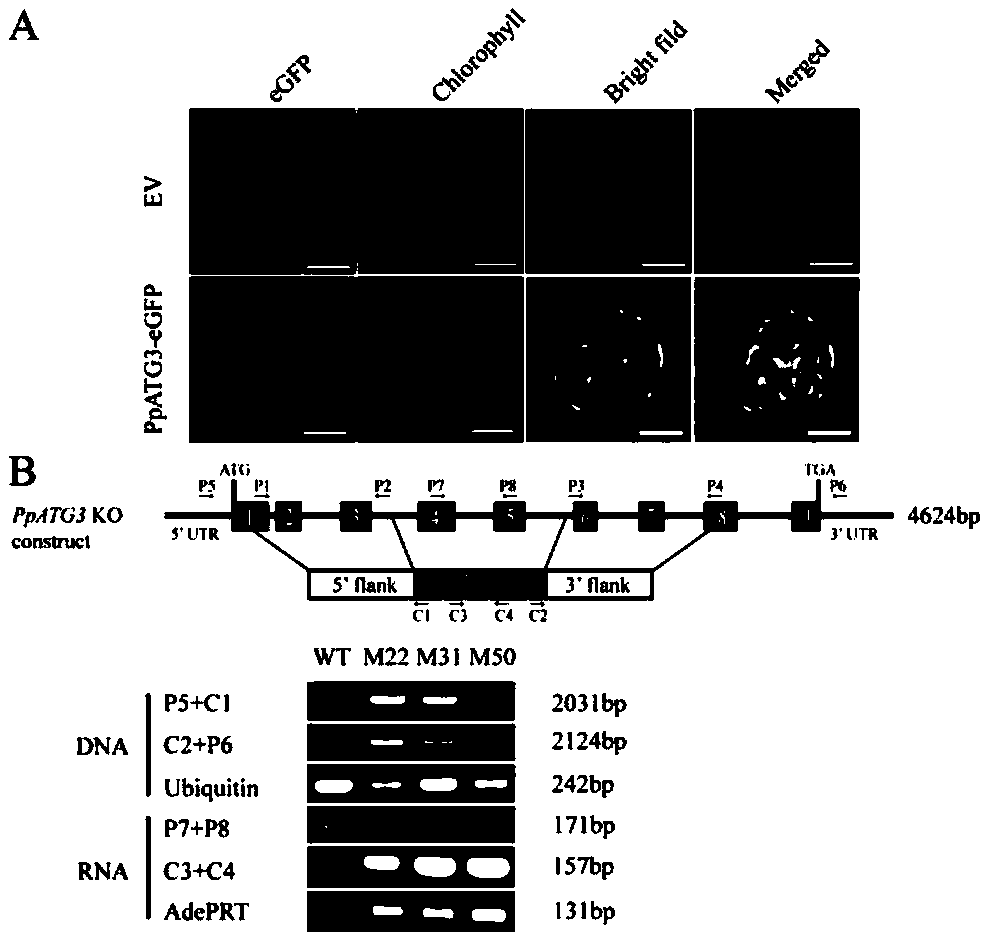
[0023] Transient expression and stable transformation of Physcomitrella patens
[0024] BCD medium was used to cultivate moss, and the protonema material was polished and subcultured for 5-6 days, and the photoperiod of 16h light / 8h dark, 500μmol·m -2 ·s -1 Light intensity, cultured at 25°C.
[0025] gene amplification
[0026] Phanta Max Super-Fidelity DNA Polymerase amplification system: Take the 50μL system as an example, 2×Phanta Max Buffer 25μL, dNTP Mix (10mM each) 1μL, DNA / cDNA 50-400ng / 1-5μL, ForwardPrimer (10μM) 2μL, Reverse Primer (10μM) 2μL, Phanta Max Super-FidelityDNAPolymerase 1μL, add ddH at the end 2 0 to 50 μL. The reaction program was as follows: pre-denaturation at 95°C for 3 min; denaturation at 95°C for 15 sec, annealing at 56-72°C for 15 sec, extension at 72°C for 60 sec / kb, 35 cycles; complete extension at 72°C for 5 min.
[0027] Restriction sites and plasmid construction
[0028] Using Physcomitrella patens cDNA as a template, using primers PpATG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com