Method for detecting differential methylation region of plant genomes
A plant genome and detection method technology, applied in the field of detection of differentially methylated regions of the plant genome, can solve the problems of unclear relationship and mechanism of plant response to drought, time-consuming detection and analysis process, unfavorable rapid analysis and detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] 1. Cultivation and drought treatment of Physcomitrella patens
[0059] Use BCDAT medium to cultivate Physcomitrella patens, get the protocelium material to polish and subculture, and light it for 14 hours (light intensity 500 μmol·m -2 ·s -1 ) / 10 hours of darkness, cultured in a culture room at 25°C for 40 days. The plant material is removed and dried and rehydrated. After about 2 hours of drying treatment, when the water loss reaches about 80%, take a sample. BCDAT medium: 0.1% TES, 1% stock solution B, 1% stock solution D, 1% stock solution C, 5mmol / L ammonium tartrate, 1mmol / L CaCl2 2H 2 O, 0.8% agar powder (solid). After autoclaving, store at room temperature.
[0060] 2. Genomic DNA extraction
[0061] The control group (M01) and the drought treatment (M02) were selected as samples, and genomic DNA was extracted by CTAB method. The quality of the extracted genomic DNA was tested by Nanodrop-2000 and agarose gel electrophoresis. CTAB extract: 2% cetyltriethy...
Embodiment 2
[0070] The methylation information of a single cytosine site on all chromosomes in the whole genome of the control group (M01) and the treatment group (M02) obtained in Example 1 is stored in a python dictionary. In order to quickly find the methylation information of a certain region on the chromosome later, the storage time is segmented (50 bp for each region) to store the information of the cytosine site. The specific form of the dictionary is {chr:{binOrder:{type:[support,unsupport,total]}}}, such as {chr1:{2:{CG:1580,265,25}}}, which means: in the first In the chromosome, in the second region (50bp is a region, starting from 1), there are a total of 25 CG-type cytosine sites, and the total number of reads that support methylation at these 25 sites is 1580, and the number of reads that do not support methylation The total number of base-based reads was 265.
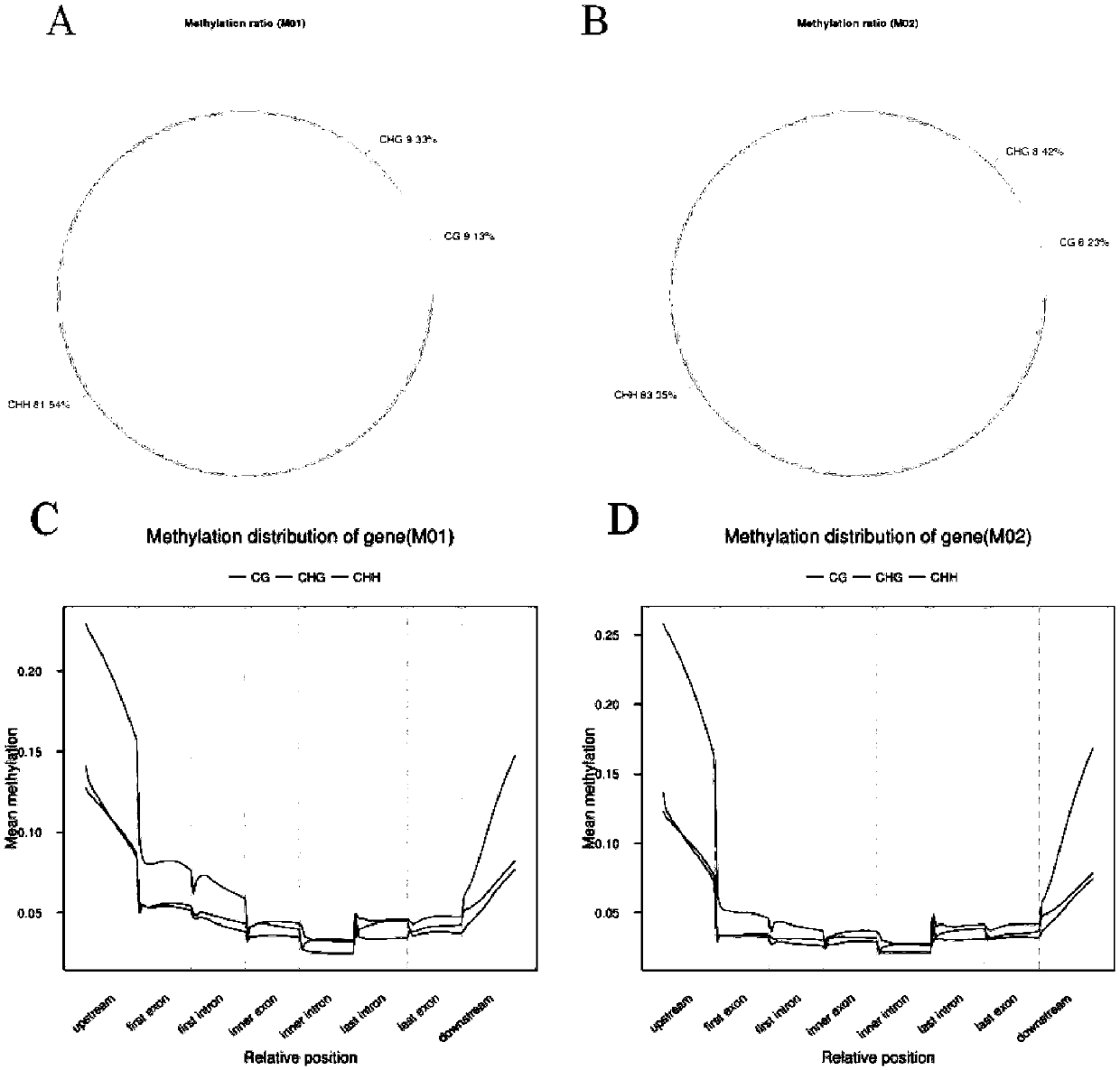
[0071] Compare the methylation information of a single cytosine site on all chromosomes in the whole genome of the...
Embodiment 4
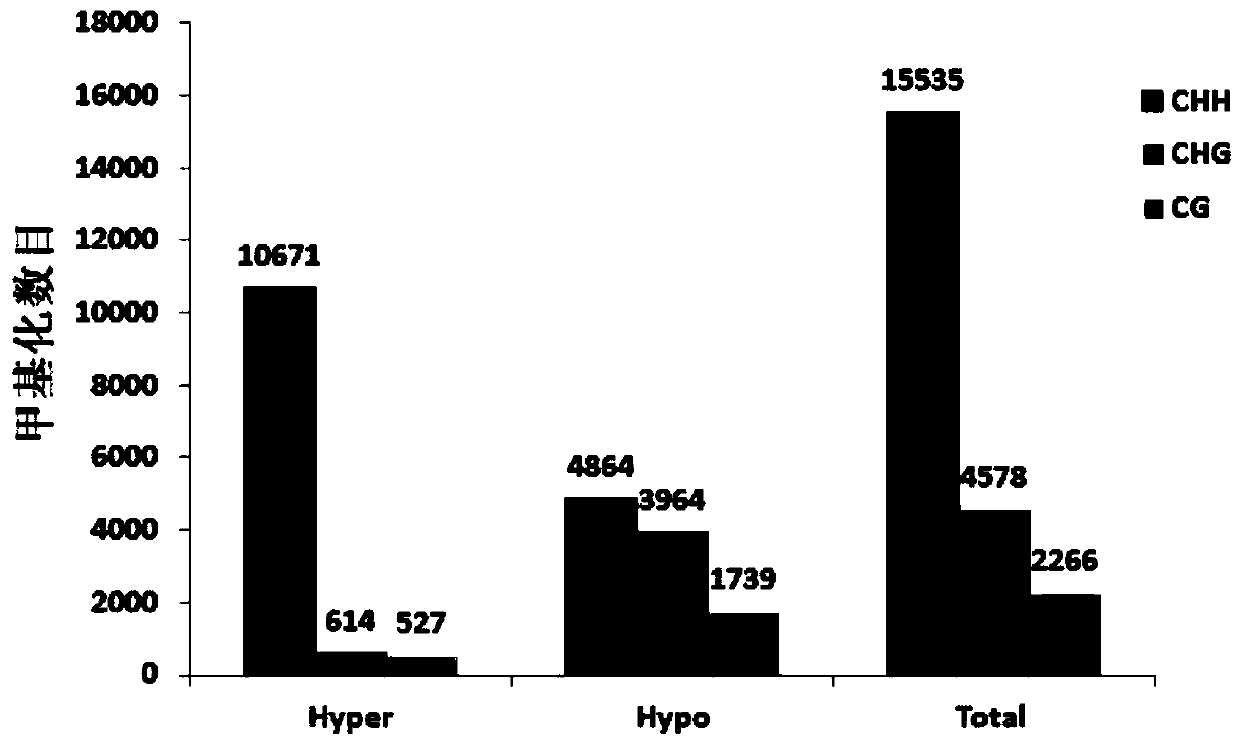
[0077] The present invention uses online databases to analyze biological processes, see the websites http: / / wego.genomics.org.cn / and http: / / kobas.cbi.pku.edu.cn / for details. The main biological processes involved in genes regulated by methylation under drought treatment were obtained through analysis (see Figure 4 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com