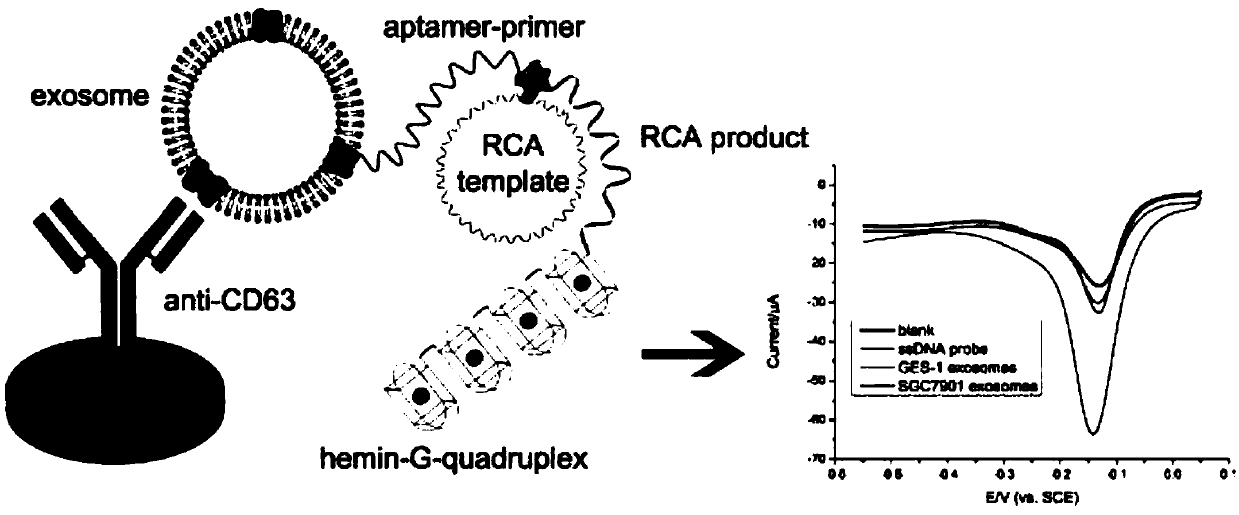
Exosome detecting method based on adapter and rolling ring amplification
A technology of rolling circle amplification and detection method, which is applied in measuring devices, material analysis through electromagnetic means, instruments, etc., can solve the problems of low sensitivity, high background signal, detection interference, etc., and achieve the effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Preparation of circular templates and rolling circle amplification products.
[0032] 1. Hybridize the phosphorylated padlock probe (100 μM) and 1 μL ligation probe (100 μM) in T4 ligase buffer; add 60 U of T4 DNA ligase, and incubate the mixture overnight at 16°C; keep T4 ligase at 65°C Inactivate for 15 minutes, add exonuclease I 20U, exonuclease III 50U to digest the remaining single-stranded DNA and double-stranded DNA; the enzyme is denatured by heating at 85°C for 10 minutes.
[0033] The sequence of the padlock probe and connection probe is as follows:
[0034] Padlock Probe:
[0035] 5’-TAGCACGGACATTTTCCCAACCCGCCCTACCCCTTTTTTTTTTCCCAACCCGCCCTACCCTTTTGTAACTGTTTCCTTC-3’
[0036] Ligation probe: 5'-TGTCCGTGCTAGAAGGAAACAGTTAC-3'
[0037] 2. Add 1μL phi29DNA polymerase buffer, 0.5μL 10mM dNTPs, 0.2μL 0.1mg / mL BSA, 10U phi29DNA polymerase, 10μL 100nM aptamer-primer probe and 10μL 120nM circular template, incubate the electrode at 37℃ 2 hours.
[0038] ...
Embodiment 2
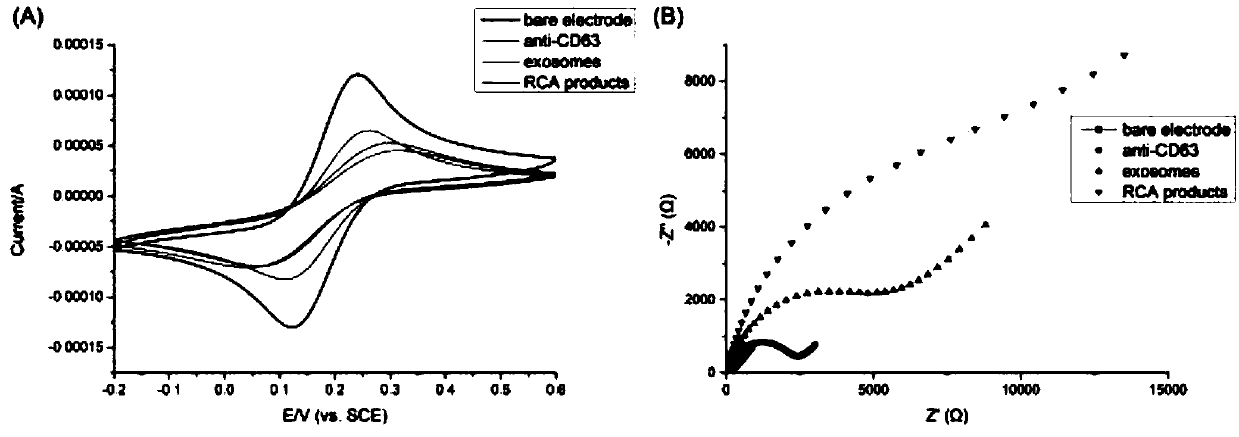
[0041] Example 2: Modifying antibodies on the surface of gold electrodes and using them to capture gastric cancer exosomes and start rolling circle amplification.
[0042] 1. The gold electrode was polished with 0.05mm alumina slurry, rinsed with ultrapure water, and dried at room temperature; the pretreated gold electrode was soaked in PBS solution containing 15mM cysteamine overnight to generate the self-activation of cysteamine. Assemble the monolayer; soak the gold electrode in 5% glutaraldehyde aqueous solution and react at 37°C for 6 hours; incubate the gold electrode with 10 μL 20 μg / mL anti-CD63 antibody at 37°C for 1 hour; Incubate in BSA solution for 1 hour at 37°C, rinse with ultrapure water.
[0043] 2. Incubate the antibody-modified electrode with the gastric cancer exosome solution resuspended in PBS for 1 hour at 37°C, and wash the electrode with PBS solution; modify 10 μL of 100 nM aptamer-primer probe to the surface of the electrode, and incubate at 37°C 1 ho...
Embodiment 3
[0045] Embodiment 3: Electrode working process
[0046] 1. Prepare the gold electrode of the modified antibody: (1) the gold electrode is polished with 0.05mm alumina slurry, the electrode is rinsed with ultrapure water, and dried at room temperature; Soak in PBS solution overnight to produce a self-assembled monolayer of cysteamine; (3) soak the gold electrode in 5% glutaraldehyde aqueous solution and react at 37°C for 6 hours; (4) mix the gold electrode with 10 μL 20 μg / mL The anti-CD63 antibody was incubated at 37°C for 1 hour; (5) The electrode was soaked in 5 mg / mL BSA solution, incubated at 37°C for 1 hour, and rinsed with ultrapure water.
[0047] 2. Circular template preparation: (1) Phosphorylated padlock probe (100 μM) and 1 μL ligation probe (100 μM) were hybridized in T4 ligase buffer; (2) 60 U T4 DNA ligase was added, and the mixture was incubated at 16 Incubate overnight at ℃; (3) Keep T4 ligase at 65℃ for 15 minutes to inactivate, add exonuclease I 20U, exonucl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com