Gene mutation detection method based on selective elimination of wild strand background interference
A detection method and single-chain technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of high price of instruments and reagents, insufficient sensitivity, long detection period, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1
[0053] In this embodiment, two different target gene DNA sequences were selected, one is the sequence where the point mutation (L858R) at codon 858 of exon 21 of the human epidermal growth factor receptor (EGFR) is located; the other is the sequence of V - the sequence of the point mutation (G13D) at codon 13 of exon 2 of Ki-ras2Kirsten rat sarcoma virus oncogene (KRAS).
[0054] Table 1. Sequences (5'-3') of nucleic acid strands used in this example
[0055]
[0056]
[0057]
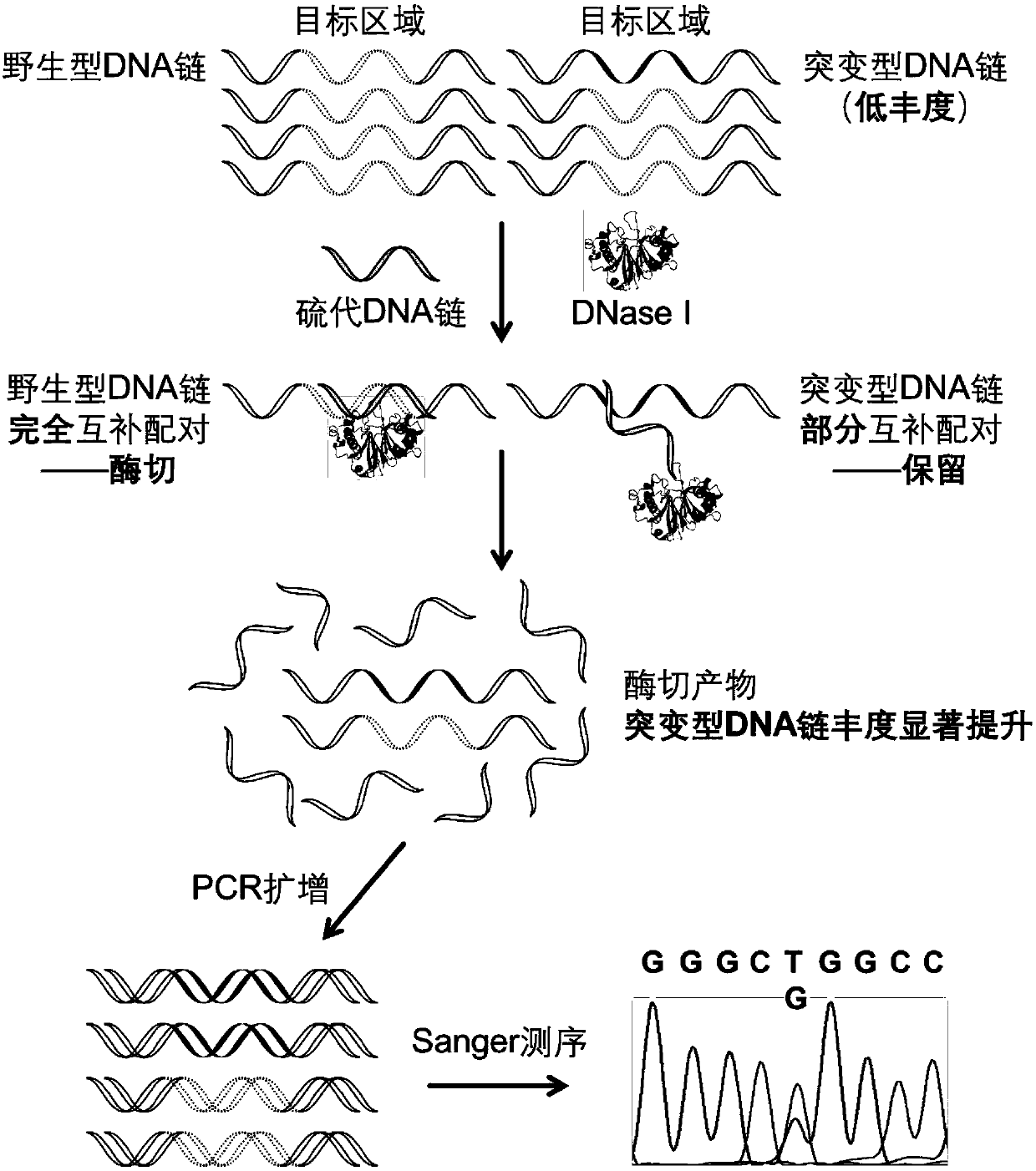
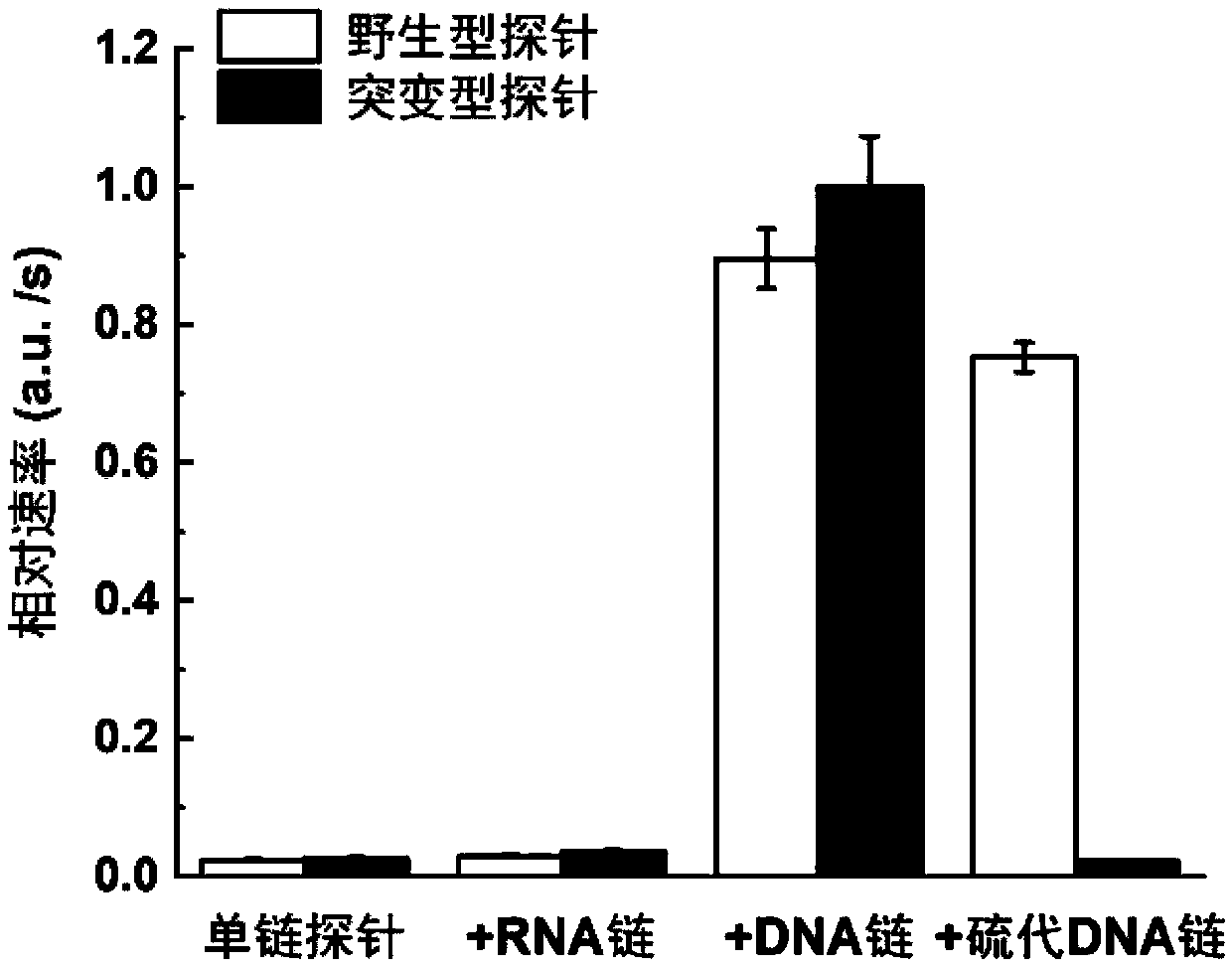
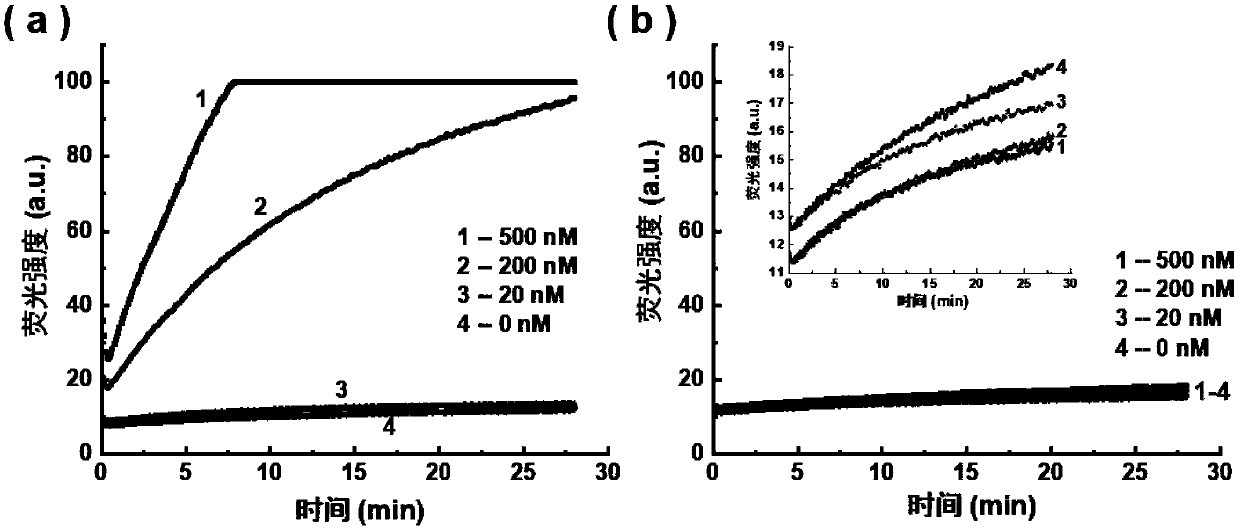
[0058] (1) EGFR L858R sulfurated DNA strand guides DNase I to selectively cut EGFR L858R wild-type DNA strand
[0059] The sequences of the EGFR L858R wild-type long-chain DNA and the EGFRL858R mutant long-chain DNA are shown in Table 1, the underlined part is the target region, and the mutation site is indicated in bold. For this target region, design the corresponding EGFR L858R sulfide DNA strand sequence as: 5'-AAAAAAAAAAAA GGGCTGGCCAA CGCAGATA-3', wherein the phosp...
Embodiment 2
[0082] Example 2
[0083] The sequences of wild-type DNA long chain, mutant DNA long chain, thio-DNA chain and RNA closed chain of EGFR L858R used in this example are listed in Table 1. The sequences of the PCR amplification primers used are:
[0084] EGFR L858R forward primer: 5'-TTCTTTTCTCTTCCGCACC-3' (5'-OH end) (SEQ ID No: 21)
[0085] EGFR L858R phosphorylation reverse primer: 5'-PO 4 -TACTTGGAGGACCGTCG-3' (5' terminal phosphorylation tag) (SEQ ID No: 22)
[0086] The experimental operation steps are as follows:
[0087] 1) Genomic DNA was digested with Shearase enzyme: Mix 1 mg of genomic DNA and 1.5 μL Shearase enzyme in 20 μL buffer, the buffer composition was 10 mmol / L Tris-HCl, 25 mmol / L MgCl 2 , 1mmol / LDTT, pH 7.5@25°C, incubate at 42°C for 15min, then heat inactivate the Shearase enzyme.
[0088] 2) PCR amplification of genomic DNA and standard DNA: 1.5 μL of the product solution in step 1) or 0.25 amol DNA standard, 20 pmol forward primer, 20 pmol reverse pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com