Carbonyl reductase ChKRED20 mutant and use thereof
A reductase and mutant technology, applied in the field of genetic engineering and enzyme engineering, can solve the problems of low catalytic ability of unnatural substrates, natural enzymes cannot adapt to industrial production conditions, etc., achieve good industrial application prospects, shorten reaction cycle, space-time The effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Error-prone PCR (error-prone PCR) method constructs carbonyl reductase random mutation library
[0035] use The II Random Mutagenesis kit randomly mutates the carbonyl reductase ChKRED20 gene.
[0036] The primers used were: T7: 5′–TAATACGACTCACTATAGGG–3′
[0037] T7ter: 5′–TGCTAGTTATTGCTCAGCGG–3′
[0038] The reaction conditions were: pre-denaturation at 95°C for 2 minutes, denaturation at 95°C for 30s, annealing at 55°C for 30s and extension at 72°C for 90s, a total of 25 cycles. After electrophoresis, the gene fragments were recovered with a gel recovery kit.
[0039] After the recovered fragment was digested with EcoR I and Sal I, it was ligated with the pET 28a (+) vector (kanamycin resistance gene) that had undergone the same digestion. The reaction conditions were: vector and fragment in molar ratio Mix at a ratio of 1:3, add 400 units of T4 DNA ligase, and ligate overnight at 16°C. The electric shock method was transferred into Escherichia coli DH...
Embodiment 2
[0040] Example 2 Screening of carbonyl reductase ChKRED20 mutant library
[0041] The mutant library clones in Example 1 were collected and the plasmids were extracted, transformed into Escherichia coli expression strain BL21-DE3, spread on LB plates containing kanamycin, and cultured for 12 hours. Single clones were picked and placed in a 96-well plate, each well contained 200 μL TB medium (containing 50 μg / mL kanamycin, 0.5 mM IPTG), 30° C., 180 rpm, and cultured with shaking for 18 hours. Use a 96-well plate replicator to replicate each single clone on an LB solid medium plate, culture at 37°C for 12 hours, and store in a refrigerator at 4°C. Centrifuge the 96-well plate after bacterial cell-induced expression at 4000 rpm for 10 min, discard the supernatant, add 200 μL of lysis buffer to each well to resuspend the cells (preparation of lysis buffer: 0.1 M, potassium phosphate buffer with pH 8.0, 10mg / mL lysozyme, 1μg / mL DNase I, 10mM MgCl 2 ). After placing the 96-well p...
Embodiment 3
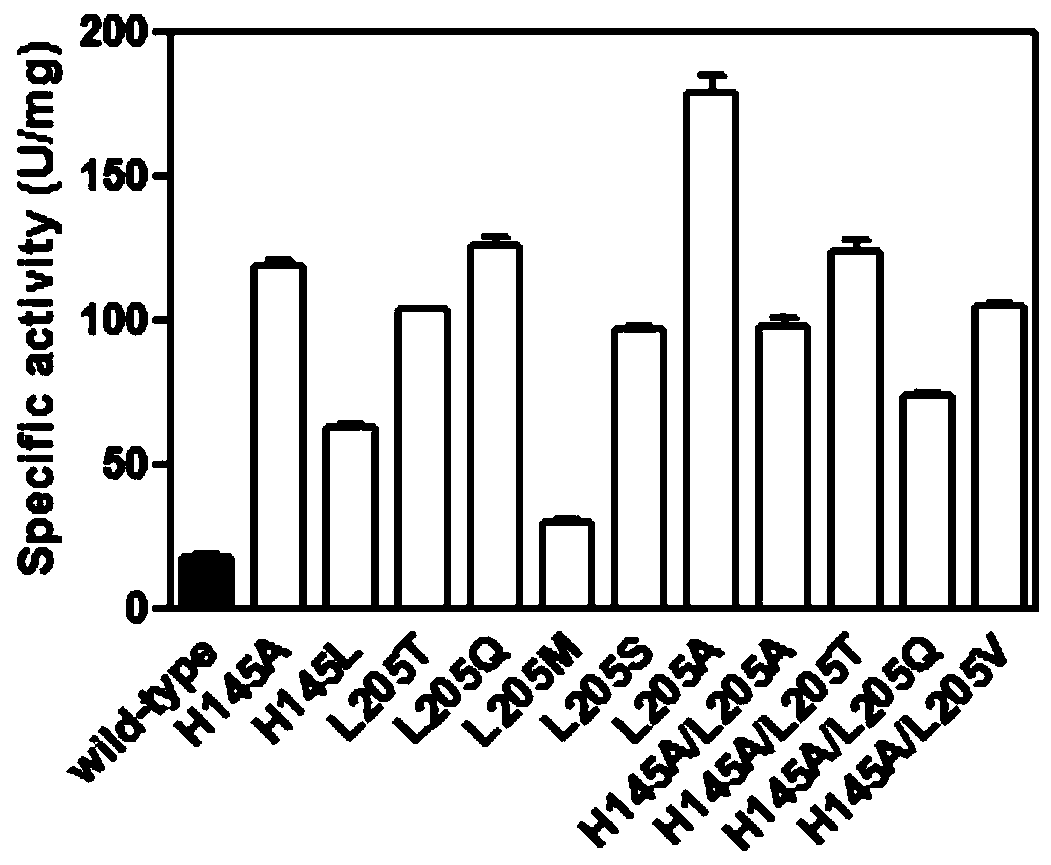
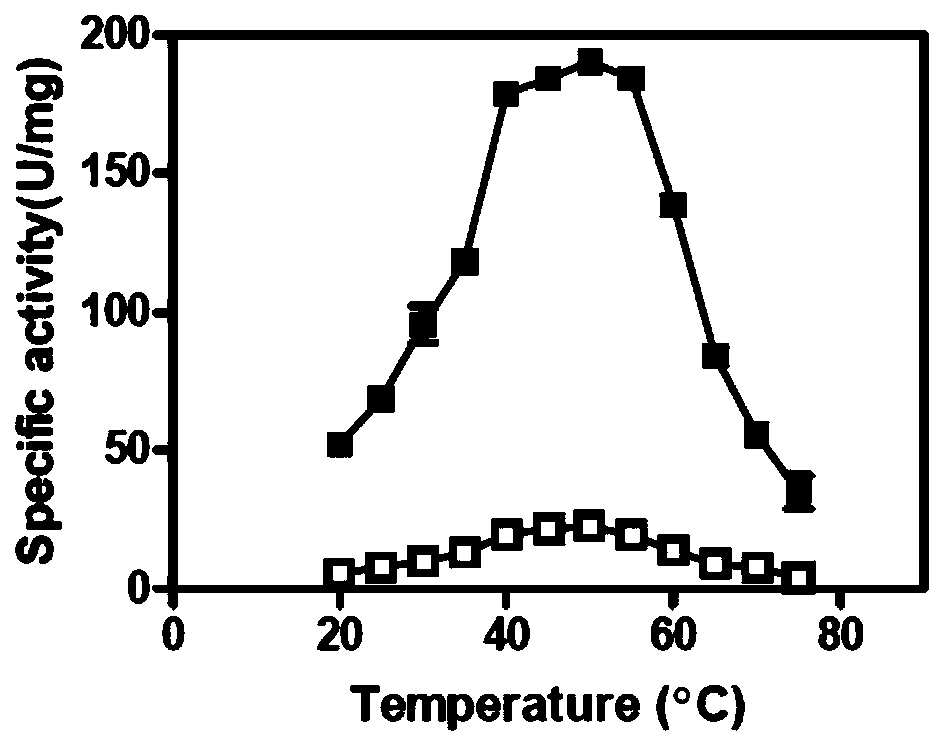
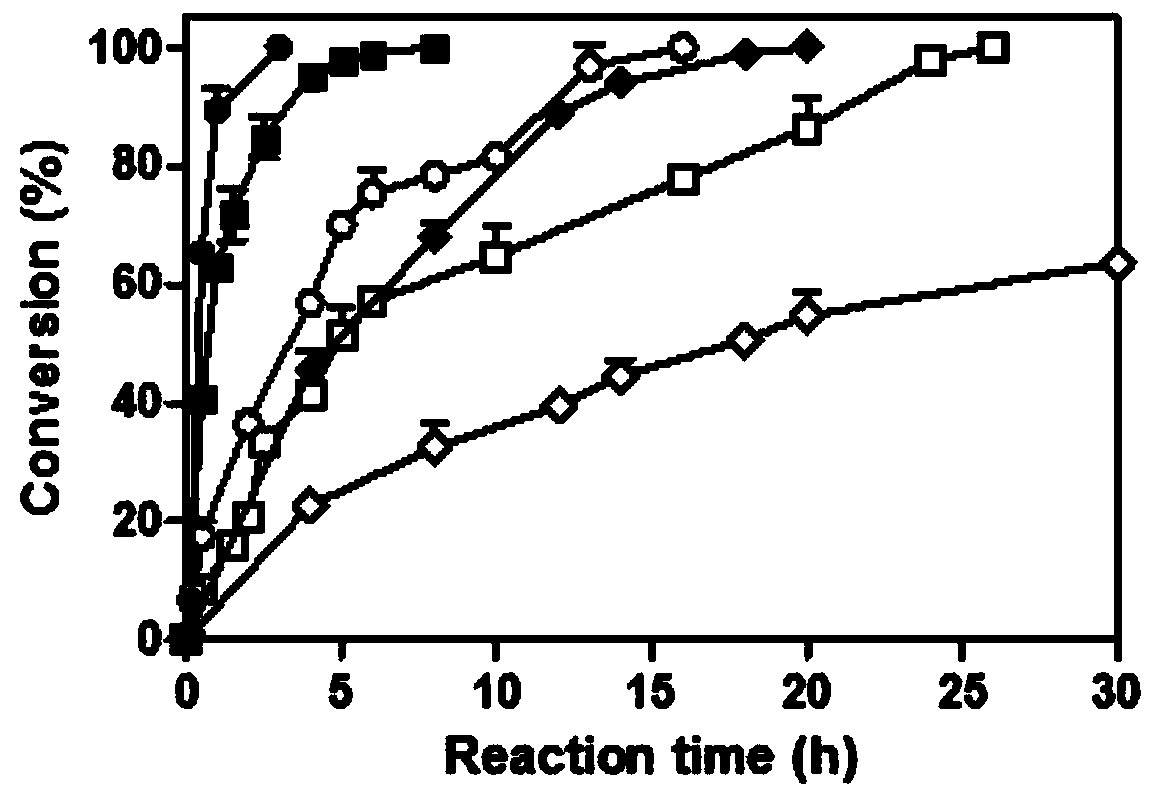
[0042] The mensuration of embodiment 3 parent and mutant specific activity
[0043] 3.1 Preparation of pure enzyme solution
[0044] The parent carbonyl reductase ChKRED20 and the mutants H145L, L205M were purified by nickel column affinity chromatography (Bio-Rad).
[0045] Pick a single clone into LB (containing kanamycin 50 μg / mL) medium, culture overnight at 37 ° C, transfer to TB (containing kanamycin 50 μg / mL) medium with 1% inoculum size, 37 Cultivate at ℃ for 3h, add 0.5mMIPTG for induction, and continue to culture at 30°C for 18h. The bacterial cells were collected by centrifugation at 4°C and 6000 rpm. Resuspended in BufferA (50mM sodium phosphate buffer, pH 8.0, 300mMNaCl, 10mM imidazole), crushed by a homogenizer, centrifuged at 13000rpm, 4°C for 20min, and added the supernatant to the column material equilibrated with Buffer A, Mix gently for 30 min, wash the impurity protein with Buffer A containing 20mM imidazole, then elute the target protein with buffer A c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com