Kit for aptamer screening and use and detection method thereof
A nucleic acid aptamer and kit technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA preparation, etc., can solve the problems of many steps, low success rate, long screening period, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] A kit for nucleic acid aptamer screening, the raw materials of which include: 1 magnet of 1 cm × 2 cm × 3 cm, 10 7 2mL streptavidin-modified magnetic beads per μL, 3 tubes of 1.3nmol / tube (dry powder) nucleic acid library, 1 tube of 5nmo / tube (dry powder) short sequence, 30 parts of 1mL PCR master mix, 150mL ePCR microbeads 1 part of droplet forming oil, 20 parts of 300 μL fluorescent quantitative PCR master mix, 1 part of 100 μL molecular weight standard sample;
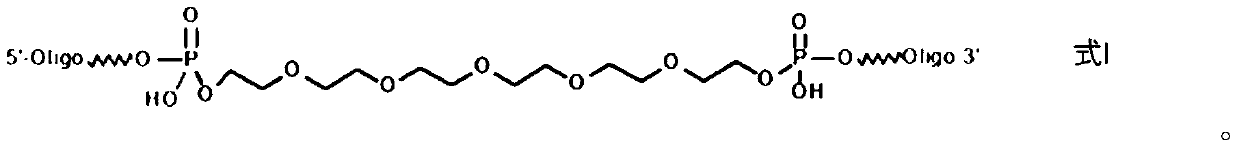
[0073] Wherein, the sequence of the nucleic acid library is 5'-ATTGGCACTCCACGCATAGG-40N-CCTATGCGTGCTACCGTGAA-3', wherein 40N represents a sequence formed by connecting 40 arbitrary nucleotide bases;
[0074] The sequence of the short sequence is: 5'-CCTATGCGTGGAGTGCCAAT-biotin-3';
[0075] The raw materials of the PCR master mix include: 100 μM FAM modified nucleic acid library forward primer 5 μL, 100 μM polyA modified nucleic acid library reverse primer 5 μL, 10 mM dNTP 20 μL, 5 U / μL DNA polymerase 4 μL, 1...
Embodiment 2
[0081] The nucleic acid aptamer screening of Example 2 was carried out with the reagents in Example 1.
[0082] A method for using a kit for nucleic acid aptamer screening, comprising the steps of:
[0083] S1. Immobilizing the nucleic acid library on magnetic beads:
[0084] Take the nucleic acid library and short sequences and use DPBS buffer as a solvent to prepare solution A. Solution A contains a nucleic acid library of Mnmol. Take solution A and pair it with a PCR machine to obtain solution B;
[0085]Wherein, during the first round of screening, the concentration of the nucleic acid library in solution A was 5 μM, and the concentration of the short sequence was 10 μM; from the second round of screening, the concentration of the nucleic acid library in solution A was 350 nM, and the concentration of the short sequence was 700 nM; The instrument pairing procedure is to keep at 95°C for 10 minutes, cool down to 60°C at a rate of 0.1°C / s, hold for 1 minute, and then cool d...
Embodiment 3
[0110] The detection in Example 3 was carried out with the fluorescent quantitative PCR master mix in Example 1.
[0111] Fluorescent quantitative PCR detection method described in embodiment 2 is:
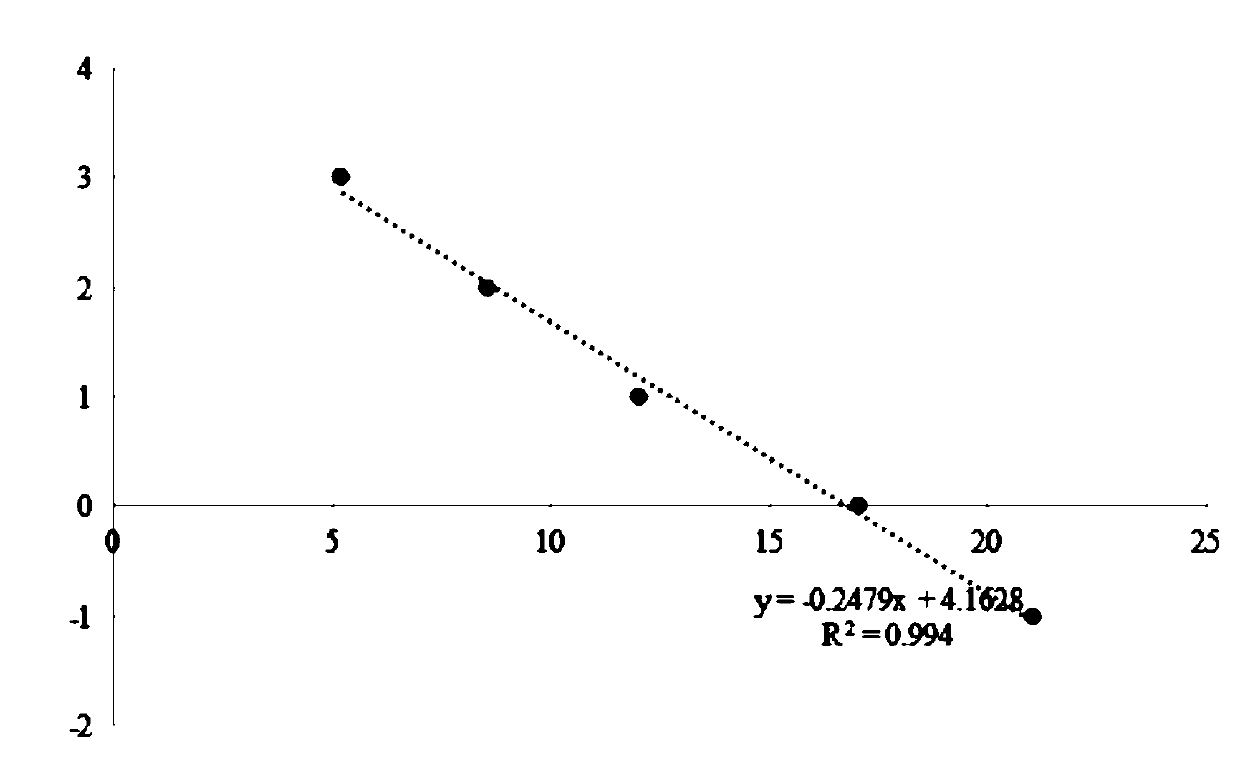
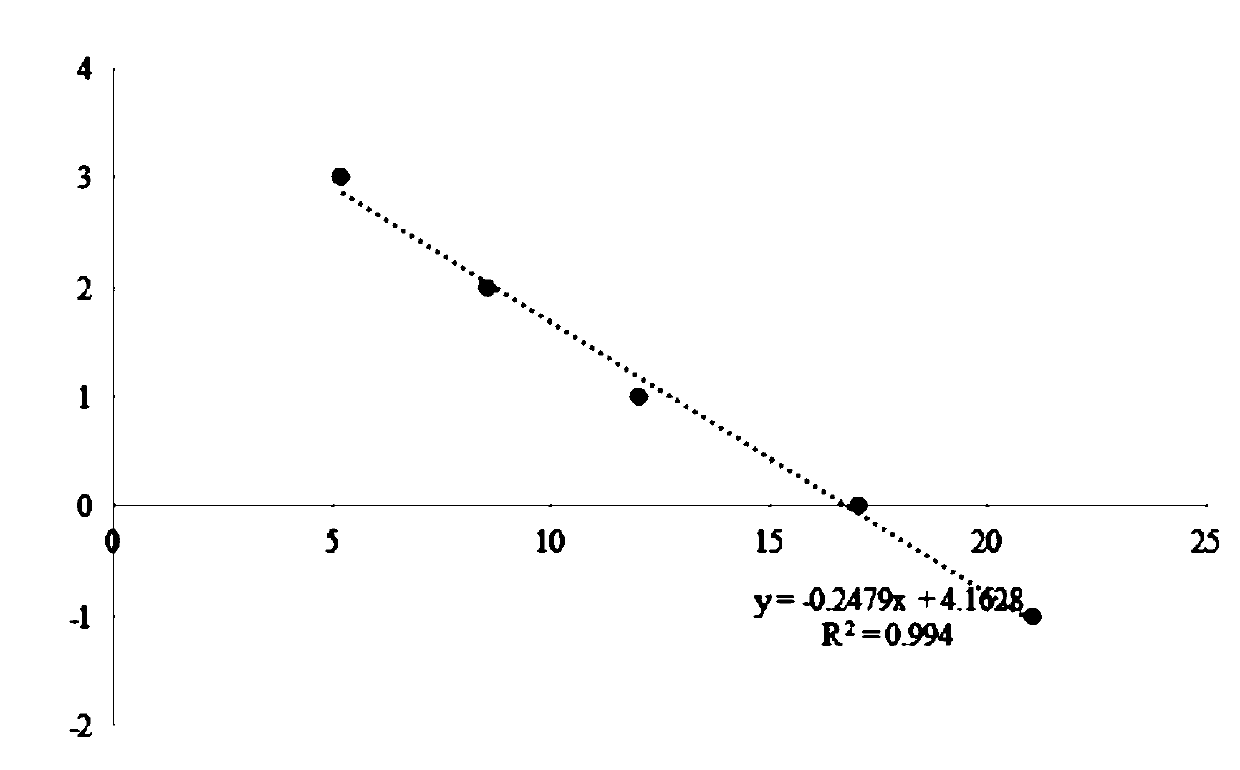
[0112] Draw a standard curve: take the nucleic acid library described in Example 1 and add it to the fluorescent quantitative PCR premix described in Example 1 to prepare standard curve solutions with different nucleic acid library concentrations, detect the Cq values of each standard curve solution by fluorescent quantitative PCR, and use each standard The lg value of the nucleic acid library concentration of the curve solution is the ordinate, and the Cq value of each standard curve solution is the abscissa to draw a standard curve to obtain a regression equation.
[0113] The values of the standard curve are shown in the table below:
[0114] X axis (Cq value) Y axis (log value of nucleic acid library concentration) Nucleic acid library concentration (pM) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com