Preparation method of Escherichia coli inner membrane vesicles endogenously highly expressing miRNA and its application in the preparation of antitumor drugs
A technology of Escherichia coli and membrane vesicles, which is applied in the direction of anti-tumor drugs, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems that have not been seen in the application of drugs loaded in inner membrane vesicles, and achieve growth inhibition, The effect of low production cost and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Preparation of Escherichia coli endogenously expressing miRNA in large quantities
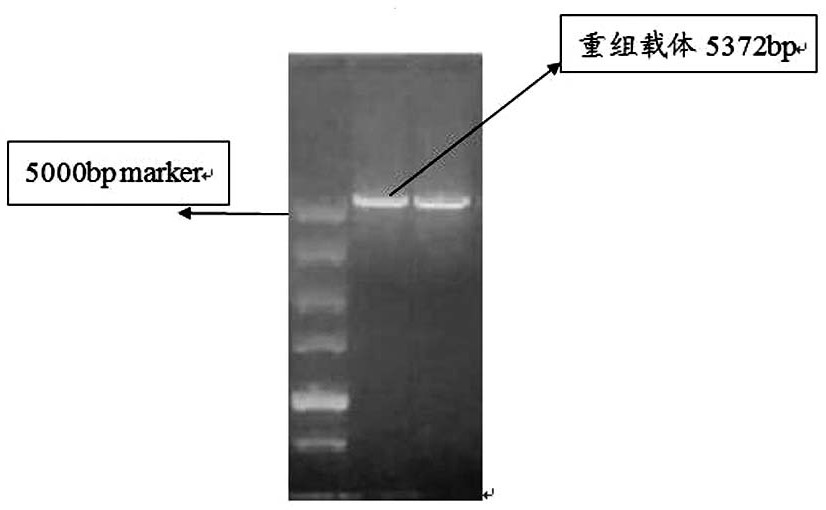
[0035] Step 1, design the primer sequence of pET-31b(+) and PCR amplify the whole gene sequence of pET-31b(+)
[0036] (1) Design the primer sequence of pET-31b(+) vector
[0037] The sequence is as follows:
[0038] Upstream primer: 5'-AAGAATTCAAGCTTATTCTCTTCTTAAAGTTAAACAAAATTATT-3';
[0039] Downstream primer: 3'-TTGGATCCGTCGAC GCAATAACTAGCATAACCCCTTG-5'.
[0040] (2) Extract the plasmid of K12 Escherichia coli as a template for PCR amplification of pET-31b(+) vector
[0041] a. Take out the K12 Escherichia coli strain preserved in 20% glycerol from the -80 low-temperature refrigerator, wait for it to melt, and streak it on a solid LB plate containing 100 mg / ml ampicillin in an ultra-clean workbench, and invert at 37°C Cultivate for 12-16 hours;
[0042] b. Pick a single clone in 5ml LB liquid medium containing 100mg / ml ampicillin, and shake and culture at 37°C and 220rm...
Embodiment 2
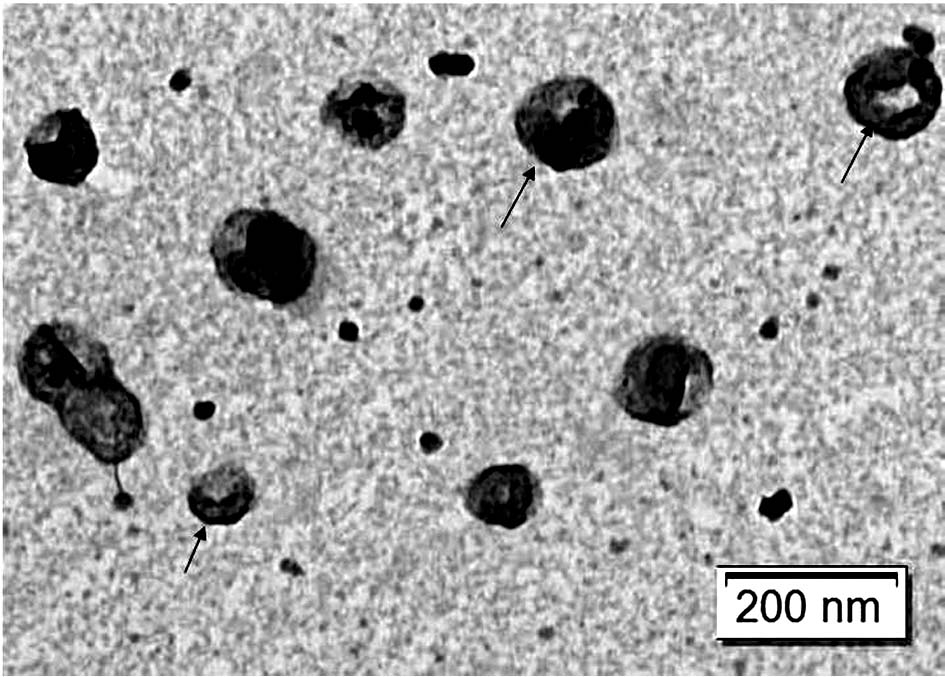
[0115] Example 2 Preparation of Escherichia coli inner membrane vesicles
[0116] 1. Use lysozyme to remove the outer cell membrane of E. coli to obtain E. coli protoplasts
[0117] a. Prepare 50mM Tris-HCl (pH 8.0), 50mM Glucose, and 1mM EDTA solution respectively as a buffer solution, and prepare lysozyme with a concentration of 4mg / ml;
[0118] b. Add lysozyme to the collected Escherichia coli cells (prepared in Example 1), react in a 37°C water bath for 20 minutes, and remove toxins such as the outer membrane of the cell and lipopolysaccharide on it;
[0119] c. Take out a little bacterial liquid and observe and detect under a microscope until completely de-walled protoplasts are obtained.
[0120] 2. Use a polycarbonate membrane to filter E. coli protoplasts to obtain nanovesicles in the inner membrane of E. coli protoplasts
[0121] a. Install a polycarbonate membrane with a pore size of 12um on a filter with a size of 47mm to filter E. coli protoplasts;
[0122] b. I...
Embodiment 3
[0125] Example 3 Fluorescent quantitative PCR detection of miR-34a in Escherichia coli inner membrane vesicles prepared in Example 2
[0126] a. Use the method provided in Example 2 to isolate E. coli inner membrane vesicles, resuspend in 100ul PBS, use 500ul RL Solution to fully lyse the bacteria, add 40ul chloroform and mix well, centrifuge at 12000rpm for 10min, and carefully pipette 200ul of the supernatant to fresh Add 400ul of absolute ethanol to a centrifuge tube, mix thoroughly and add to the chromatography column, centrifuge at 12000rpm for 1 minute, pour off the filtrate, wash twice with Wash Buffer, and finally use RNase-free DEPC water to elute RNA to obtain fresh RNA;
[0127] b. Use the downstream primer of miR-34a in step 2 for reverse transcription, and reverse transcribe the RNA obtained in the previous step into cDNA; RNA 3ul, 10×Buffer 2ul, reverse transcriptase 2ul, downstream primer 1ul, DEPC water 12ul, the total system is 20ul, select the reverse transc...
PUM
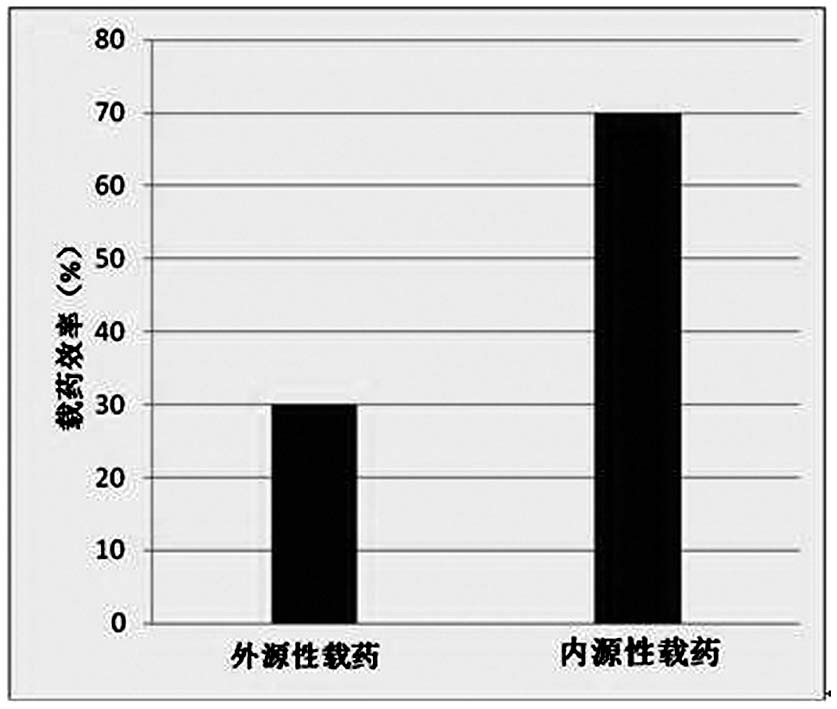
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com