Metagenome extraction method for removing host genomic DNA based on CRISPR-Cas
A metagenomic and extraction method technology, applied in the field of CRISPR technology and the reduction of host genomic DNA in metagenomic nucleic acid samples, can solve the problems of low removal efficiency, loss of nucleic acid, and high cost, and achieve high removal efficiency, simple steps, and high specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
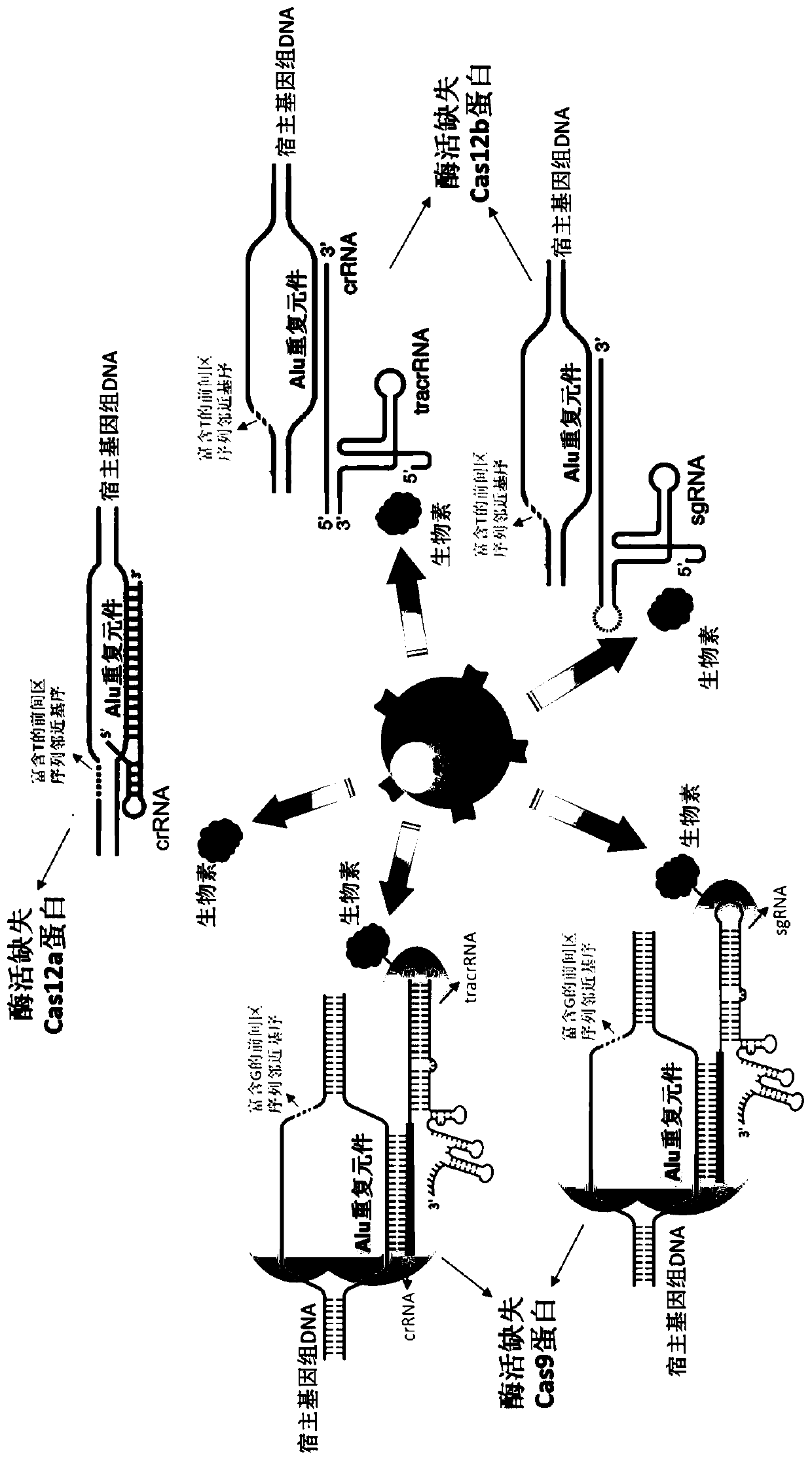
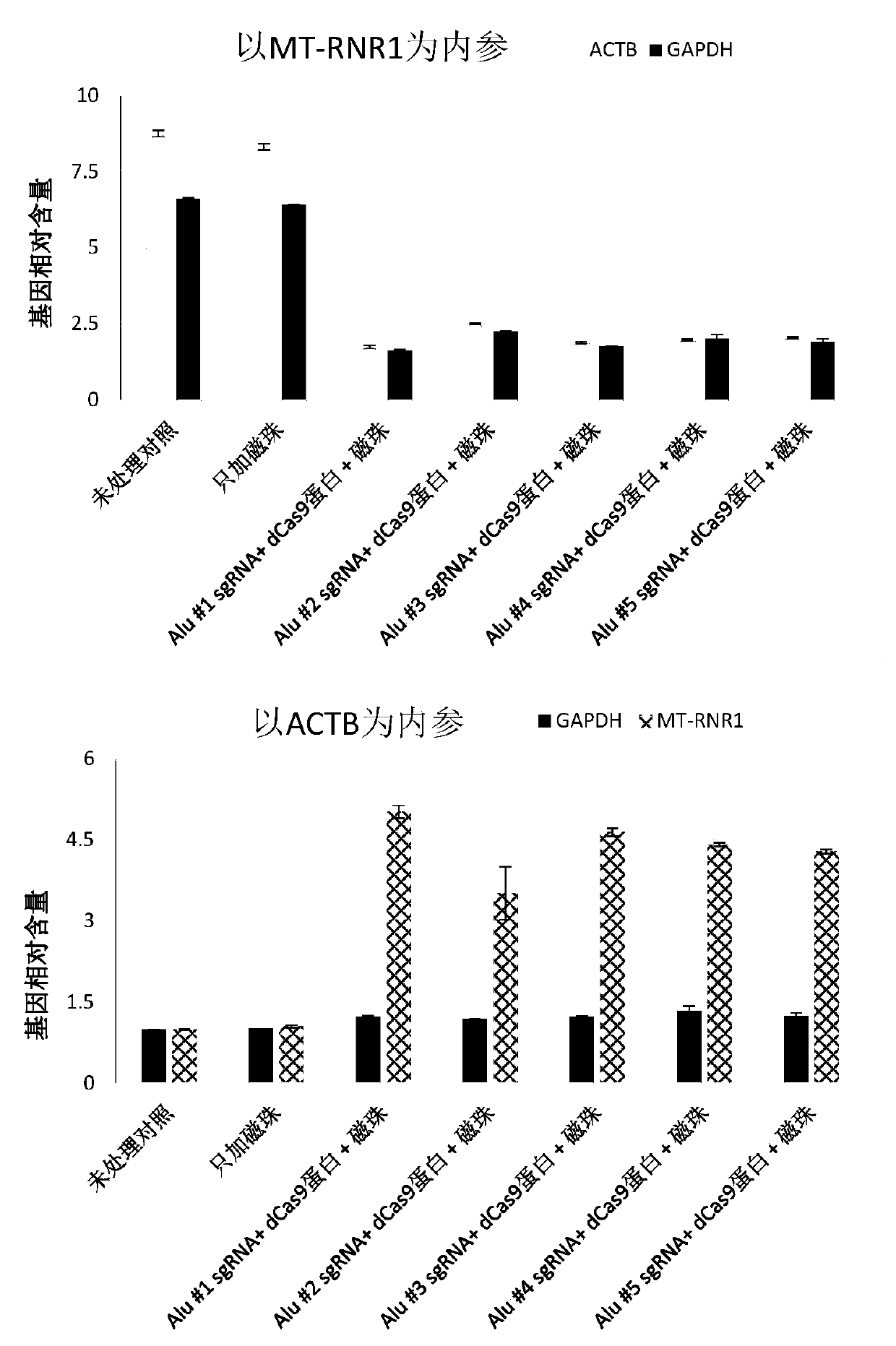
[0052] Example 1 Using SpCas9 protein with missing enzyme activity and several sgRNAs targeting Alu elements to remove human genomic DNA in nucleic acid samples extracted from whole blood
[0053] (1) Extract metagenomic nucleic acid from a 2ml whole blood sample: the whole blood sample is taken from a patient with clinically suspected pneumonia. The nucleic acid extraction kit uses QIAGEN's QIAamp DNA Blood Midi Kit (Cat. No.: 51183). Refer to its operation manual for operation. After the extraction is complete, use ThermoFisher's Qubit TM dsDNA HS Assay Kit (Cat. No.: Q32851) reagent for DNA concentration determination of the extracted nucleic acid. Dilute with nuclease-free purified water to adjust the DNA concentration to 50-200ng / μl. The extracted metagenomic nucleic acid can be stored at -20°C.
[0054] (2) Preparation of Cas9 guide RNA (sgRNA) targeting Alu element:
[0055] The guide RNA can be directly synthesized by a company such as IDT, and can also be prepared by usi...
Embodiment 2
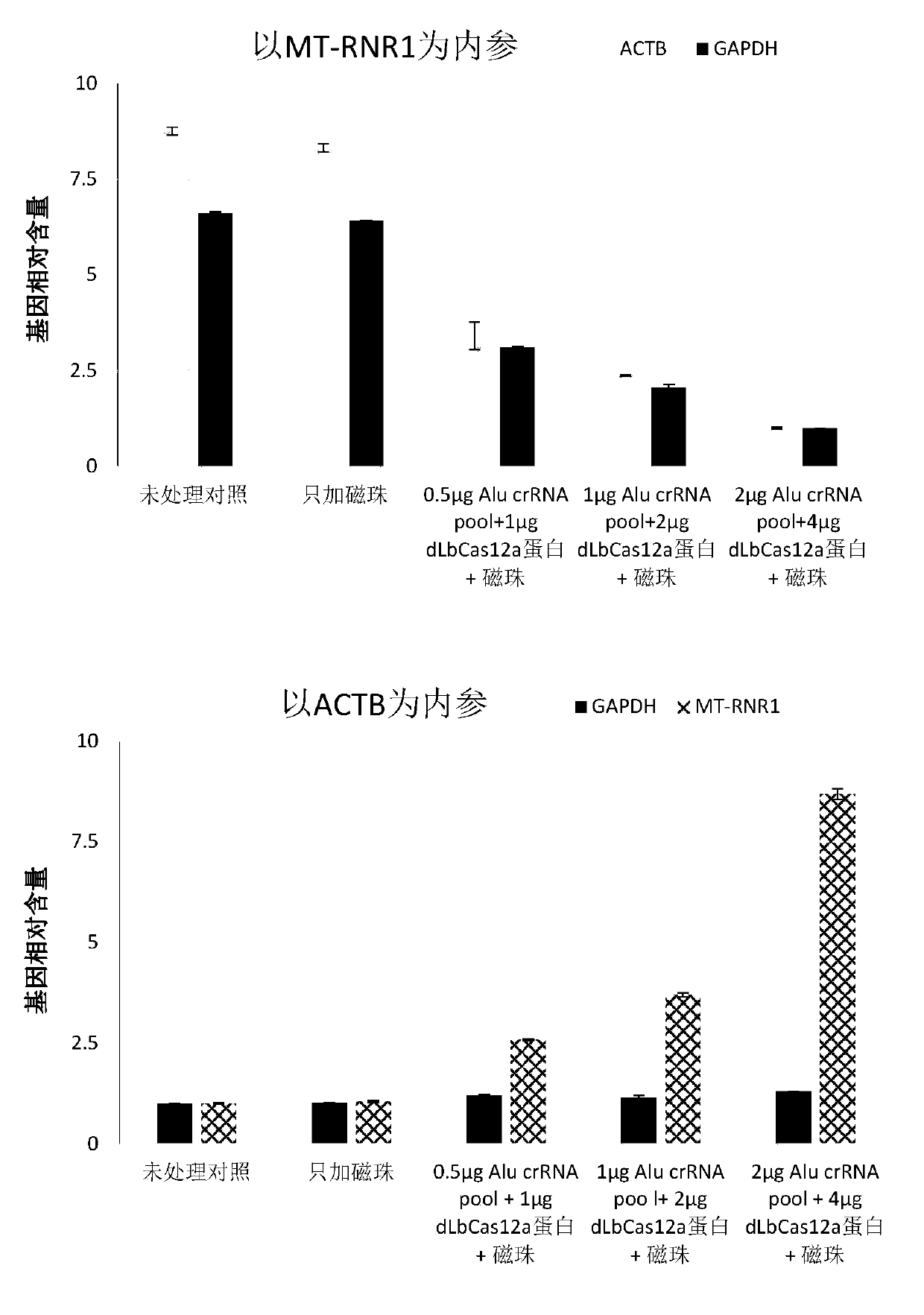
[0069] Example 2 The use of different doses of enzyme-depleted LbCas12a protein and a mixture of crRNA targeting the Alu element to remove human genomic DNA in nucleic acid samples extracted from whole blood
[0070] (1) Extract metagenomic nucleic acid from a 2ml whole blood sample: the whole blood sample is taken from a patient with clinically suspected pneumonia. The nucleic acid extraction kit uses QIAGEN's QIAamp DNA Blood Midi Kit (Cat. No.: 51183). Refer to its operation manual for operation. After the extraction is complete, use ThermoFisher's Qubit TM dsDNA HS Assay Kit (Cat. No.: Q32851) reagent for DNA concentration determination of the extracted nucleic acid. Dilute with nuclease-free purified water to adjust the DNA concentration to 50-200ng / μl. The extracted metagenomic nucleic acid can be stored at -20°C.
[0071] (2) Synthesize and order LbCas12a crRNA targeting Alu elements: crRNA can be directly synthesized by companies such as IDT. This example is designed fo...
Embodiment 3
[0083] Example 3 Using a mixture of SpCas9 (dCas9) protein with lack of enzyme activity and sgRNA targeting Alu element to remove human genomic DNA in nucleic acid samples extracted from urine, and combining with NEB Microbiome DNAEnrichment kit comparison removal efficiency
[0084] (1) Extract metagenomic nucleic acid from three 4ml urine samples: Three urine samples were taken from different patients with clinically suspected urinary tract infections. The nucleic acid extraction kit used QIAGEN’s QIAamp DNA Mini Kit (Cat. No. 51304), reference Its operation manual for operation. After the extraction is complete, use ThermoFisher's Qubit TM dsDNA HS Assay Kit (Cat. No.: Q32851) reagent for DNA concentration determination of the extracted nucleic acid. Dilute with nuclease-free purified water to adjust the DNA concentration to 50-200ng / μl. The extracted metagenomic nucleic acid can be stored at -20°C.
[0085] (2) Preparation of Cas9 guide RNA (sgRNA) targeting Alu element: T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com